3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(19):3557-3569. doi:10.7150/jca.26350 This issue Cite
Review
The Role of MicroRNAs in Hepatocellular Carcinoma
1. Shanghai Municipal Hospital of Traditional Chinese Medicine, Shanghai University of Traditional Chinese Medicine, Shanghai, 200071, P.R. China.
2. Department of Clinical Laboratory Medicine, Shanghai Municipal Hospital of Traditional Chinese Medicine, Shanghai University of Traditional Chinese Medicine, Shanghai, 200071, P.R. China.
3. Shanghai Chest Hospital, Shanghai Jiao Tong University, Shanghai, 200030, P.R. China.
# These authors contributed equally to this study.
Received 2018-3-28; Accepted 2018-7-23; Published 2018-9-8
Abstract
Hepatocellular carcinoma (HCC) is one of the most common cancers, leading to the second cancer-related death in the global. Although the treatment of HCC has greatly improved over the past few decades, the survival rate of patients is still quite low. Thus, it is urgent to explore new therapies, especially seek for more accurate biomarkers for early diagnosis, treatment and prognosis in HCC. MicroRNAs (miRNAs), small noncoding RNAs, are pivotal participants and regulators in the development and progression of HCC. Great progress has been made in the studies of miRNAs in HCC. The key regulatory mechanisms of miRNAs include proliferation, apoptosis, invasion, metastasis, epithelial-mesenchymal transition (EMT), angiogenesis, drug resistance and autophagy in HCC. And exosomal miRNAs also play important roles in proliferation, invasion, metastasis, and drug resistance in HCC by regulating gene expression in the target cells. In addition, some miRNAs, including exosomal miRNAs, can be as potential diagnostic and prediction markers in HCC. This review summarizes the latest researches development of miRNAs in HCC in recent years.
Keywords: microRNAs, hepatocellular carcinoma, exosomes, regulatory mechanism, diagnosis, prediction, marker
Introduction
Hepatocellular carcinoma (HCC) has become the second most common cause of cancer-related death worldwide [1], with approximately 782,500 new cases and 745,500 deaths occurring in the global during 2012 [2]. In the early stage of HCC, surgical resection, liver transplant, local ablation and other curative therapies can improve patient´s survival [3]. However, the 5-year recurrence rate is very high, it may reach as high as 80%-90% even the HCC patients have received potentially curative therapies [4]. It has been already advanced stage for most people when HCC was diagnosed [5]. For the advanced stage, the small molecule targeted therapeutics drugs sorafenib and regorafenib are the standard treatments that have been approved by the US Food and Drug Administration (FDA). Sorafenib is the only standard first-line systemic therapy available for advanced HCC, but the median survival was reported only 3 months [6]. Regorafenib is a second-line drug when HCC patients were progressing on sorafenib treatment, whereas, the median survival was still only 10.6 months according to a phase 3 clinical trial report [7]. Even though sorafenib and regorafenib can improve overall survival of HCC patients, it is not too long. Furthermore, the worries for drug resistance and adverse action of these drugs are rising as well. Therefore, it is urgent to explore new therapies, especially seek for more accurate markers for early diagnosis, treatment and prognosis in HCC. Nucleic acid-based drugs such as microRNAs (miRNAs) may have the promising therapeutic potential for HCC treatment. MiRNAs are pivotal participants and regulators in the development and progression of HCC. And exosomal miRNAs also play important roles in the development and progression in HCC. In addition, some miRNAs, including exosomal miRNAs, can be as diagnostic and prediction markers in HCC. In this review, we summarize the latest researches development of miRNAs in HCC in recent years.
The process of miRNA formation. At first, miRNA genes are transcribed to primary pri-miRNAs by RNA polymerase II in the nucleus. Pri-miRNAs are cleaved by the RNase III type endonuclease Drosha producing pre-miRNAs the next. After that, being transported by exportin-5 from nucleus to cytoplasm, pre-miRNAs are processed by another RNase III type endonuclease Dicer to generate a miRNA protein complex with two strands. One strand of the complex will become a mature miRNA, and then the mature miRNA is bound to RNA-mediated silencing complexes (RISC) immediately. In the RISC, the mature miRNA targets the 3'-untranslated regions of its target mRNAs to regulate translational inhibition or mRNA cleavage. The other one will be degraded.
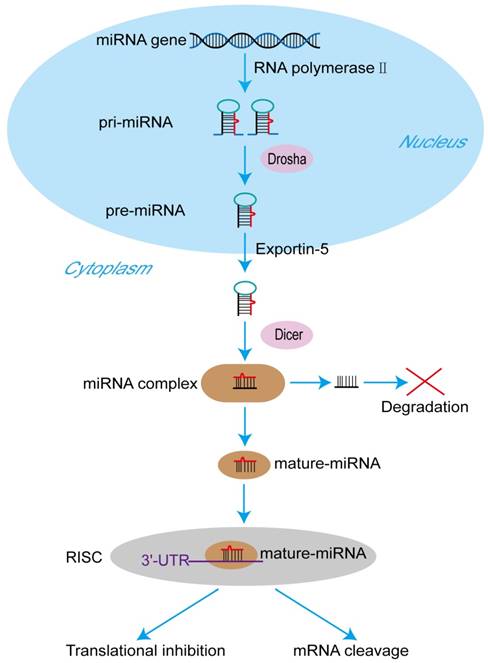
Biogenesis of miRNAs
The sequence of the human genome has been finished in 2003, and it was reported that only 20,000-25,000 genes, about 1.5% of the total human genome, can encode protein [8]. In other words, noncoding RNAs (ncRNAs), including miRNAs, long noncoding RNAs (lncRNAs), small nuclear RNAs (snRNAs) and circularRNAs (circRNAs), are the major components of the human transcriptome [9]. MiRNAs are the pivotal members of this noncoding RNA family [10]. MiRNAs, ~ 23 nucleotides in length, act as important gene regulators in animals and plants [11]. MiRNAs control the expression of their target mRNAs principally by binding to the 3'-untranslated region (3'-UTR) [12]. A mature miRNA formation goes through a series of complicated process. It was described in the Figure 1. At first, miRNA genes are transcribed to primary microRNAs (pri-miRNAs) by RNA polymerase II in the nucleus [13]. Pri-miRNAs are cleaved by the RNase III type endonuclease Drosha the next, resulting in releasing the precursor miRNAs (pre-miRNAs), which have about ~70 nucleotides and stem-loop structures [14]. After that, being transported by exportin-5 from nucleus to cytoplasm, pre-miRNAs are processed by another RNase III type endonuclease Dicer to generate a miRNA protein complex with two strands [15, 16]. One strand will become a mature miRNA, and then the mature miRNA is bound to RNA-mediated silencing complexes (RISC) immediately [17]. In the RISC, the mature miRNA targets the 3'-UTR of its target mRNAs to regulate gene posttranscriptional expression, including translational inhibition and mRNA cleavage [18]. The other one will be degraded. It has been proved that miRNAs play crucial roles in multiple biological processes by regulating gene expression, and the abnormal expression of miRNAs are related to numerous cancers and many other diseases [19].
miRNAs and HCC
The research in miRNAs and their relevant functional mechanisms of cancer will contribute to the development of the therapeutics. Thus, we summarize recent researches development in regulating miRNAs in HCC. Some miRNAs have been found to be upregulated in HCC, which can be seen in the Table 1 [20-53], and some downregulated can be seen in the Table 2 [54-146]. The key regulatory mechanisms of miRNAs in these studies include proliferation, apoptosis, invasion, metastasis, epithelial-mesenchymal transition (EMT), angiogenesis, drug resistance and autophagy in the development and progression of HCC. In addition, some miRNAs can also be as potential diagnostic and prediction markers in HCC.
Upregulated miRNAs in HCC
| MiRNA | Targets | Mechanisms | References |
|---|---|---|---|
| miR-10b | CSMD1 | Migration, invasion | [20] |
| miR-21 | CAMSAP1, DDX1, MARCKSL1 | No mentioned | [21] |
| miR-25 | RhoGDI1, TRAIL | EMT, apoptosis | [22, 23] |
| miR-32 | No mentioned | Prognostic marker | [24] |
| miR-92a | FBXW7 | Cell growth, prognostic marker | [25] |
| miR-96-5p | Caspase-9 | Apoptosis | [26] |
| miR-107 | Axin2, HMGA2, HMGCS2 | Proliferation, prognostic marker | [27-29] |
| miR-135a | FOXO1 | Migration, invasion | [30] |
| miR-155-5p | PTEN | Proliferation, apoptosis, invasion, migration | [31] |
| miR-181a | Atg5 | Autophagy | [32] |
| miR-182 | TP53INP1 | Drug resistance | [33] |
| miR-197 | CD82 | Migration, invasion | [34] |
| miR-203a-3p.1 | IL-24 | Cell growth, proliferation, metastasis | [35] |
| miR-210 | FGFRL1, YES1 | Metastasis, angiogenesis, proliferation | [36, 37] |
| miR-214-5p | WASL | Migration, invasion, EMT | [38] |
| miR-216a/217 | PTEN, SMAD7 | Drug resistance | [39] |
| miR-221 | No mentioned | Prognostic marker | [40] |
| miR-302d | TGFBR2 | Cell growth, apoptosis, migration | [41] |
| miR-331-3p | ING5 | Proliferation, apoptosis | [42] |
| miR-346 | FBXL2 | Proliferation, migration, invasion | [43] |
| miR-454 | CHD5 | Proliferation, EMT, prognostic marker | [44] |
| miR-487a | SPRED2, PIK3R1 | Proliferation, metastasis, prognostic marker | [45] |
| miR-765 | INPP4B | Proliferation | [46] |
| miR-873 | TSLC1 | Proliferation, migration, invasion | [47] |
| miR-892a | CD226 | Proliferation, invasion | [48] |
| miR-1246 | CADM1 | Migration, invasion, diagnostic and prognostic marker | [49] |
| miR-1249 | PTCH1 | Cell growth, migration, invasion | [50] |
| miR-1468 | CITED2, UPF1 | Proliferation, apoptosis | [51] |
| miR-3910 | MST1 | Cell growth, migration | [52] |
| miR-4417 | TRIM35 | Proliferation, apoptosis | [53] |
MiRNAs and proliferation and apoptosis of HCC
Cell growth, proliferation and apoptosis are the significant processes that guarantee the internal stability and balance of cell number and biological functions [14]. Cell proliferation is achieved through the cell cycle, a strictly and orderly controlled process of cell activity. The cyclin dependent kinases (CDKs) are the core regulators of the cell cycle [147]. Any cell proliferation process follows certain rules. When the cell cycle is out of control and cell unlimited proliferate, it will develop into a tumor cell [148]. Apoptosis is also called as programmed cell death [149]. Apoptosis contributes to maintain the internal balance between cell death and renewal [150]. Disorder of apoptosis is often associated with human diseases, for example, deficient apoptosis may lead to tumor [151]. In short, cell unlimited proliferation and abnormal regulation of apoptosis will promote the formation of cancer, including HCC.
Downregulated miRNAs in HCC
| MiRNA | Targets | Mechanisms | References |
|---|---|---|---|
| miR-7 | mTOR, TYRO3 | Autophagy, drug resistance | [54, 55] |
| miR-7/21/107 | Maspin | Drug resistance, prognostic marker | [56] |
| miR-26 | ULK1 | Autophagy | [57] |
| miR-29a | CLDN1 | Proliferation, migration | [58] |
| miR-30a-5p | AEG-1 | Cell growth, apoptosis | [59] |
| miR-30e | MTA1 | EMT | [60] |
| miR-31 | NDRG3 | Drug resistance | [61] |
| miR-31-5p | SP1 | Proliferation, migration, invasion | [62] |
| miR-33a | No mentioned | Prognostic marker | [63] |
| miR-33a-5p | No mentioned | Drug resistance | [64] |
| miR-33b | SALL4 | Proliferation, metastasis | [65] |
| miR-98 | EZH2 | Proliferation | [66] |
| miR-101 | Mcl-1, RAB5A, STMN1, ATG4D | Apoptosis, autophagy, diagnostic marker | [67-69] |
| miR-105-1 | NCOA1 | Diagnostic and prognostic marker | [70] |
| miR-122 | Snail1, Snail2, PKM2, DLX4 | EMT, proliferation, apoptosis, prognostic marker | [71-73] |
| miR-124-3p | MAPK14, RELA, CDK2, CDK4, SP1 | No mentioned | [74] |
| miR-126 | VEGF | Angiogenesis | [75] |
| miR-137 | EZH2 | Proliferation, invasion | [76] |
| miR-138 | Cyclin D3, SP1 | Prognosis marker, Proliferation, invasion, migration | [77, 78] |
| miR-142 | THBS4, TGF-β | Migration, invasion, cell growth, metastasis | [79, 80] |
| miR-142-3p | ATG5, ATG16L1, LDHA | Autophagy, drug resistance, proliferation | [81, 82] |
| miR-143 | TLR2 | Proliferation, invasion | [83] |
| miR-144 | ZFX | Proliferation, invasion, migration | [84] |
| miR-146a | HAb18G | Metastasis, angiogenesis | [85] |
| miR-152 | RTKN, DNMT1 | Cell growth | [86, 87] |
| miR-186 | YAP1 | Migration, invasion, proliferation | [88] |
| miR-187-3p | S100A4 | EMT | [89] |
| miR-194 | MAP4K4 | Proliferation, diagnostic and prognostic marker | [90] |
| miR-195 | Wnt3a, CBX4, FGF2, VEGFA | Proliferation, metastasis, angiogenesis | [91-93] |
| miR-199 | RGS17 | Proliferation, migration, invasion | [94] |
| miR-199a-3p | VEGFA, VEGFR1, VEGFR2, HGF, MMP2, YAP1 | Angiogenesis, proliferation, apoptosis | [95, 96] |
| miR-199a-5p | CLTC | Cell growth | [97] |
| miR-199b-5p | TGF-β1 | EMT | [98] |
| miR-200a | CXCL1, GAB1 | EMT, invasion, migration | [99, 100] |
| miR-203 | IL-1β, Snail1, Twist1 | Proliferation, metastasis | [101] |
| miR-206 | CCND1, cMET, CDK6 | Proliferation, apoptosis | [102] |
| miR-211 | SPARC | Proliferation, migration, invasion | [103] |
| miR-212 | FOXM1 | Migration, cell growth | [104] |
| miR-217 | MTDH | Proliferation, apoptosis, migration, invasion | [105] |
| miR-223 | Rab1 | Proliferation, apoptosis | [106, 107] |
| miR-296 | FGFR1 | Proliferation, apoptosis, prognostic marker | [108] |
| miR-320a | c-Myc | Proliferation, invasion | [109] |
| miR-337 | HMGA2 | Proliferation, apoptosis | [110] |
| miR-338-3p | TAZ, MACC1, β-catenin, VEGF | Proliferation, migration, angiogenesis | [111, 112] |
| miR-340 | JAK1 | Proliferation, invasion | [113] |
| miR-345 | IRF1 | Metastasis, EMT | [114] |
| miR-361-5p | VEGFA | Proliferation, invasion | [115] |
| miR-365 | ADAM10 | Proliferation, metastasis | [116] |
| miR-367-3p | MDM2 | Drug resistance | [117] |
| miR-370 | PIM1 | Cell growth, invasion | [118] |
| miR-375 | HMSN | Drug resistance | [119] |
| miR-377 | Bcl-xL | Apoptosis | [120] |
| miR-429 | RAB23 | Metastasis, EMT | [121] |
| miR-451 | IL-6R | Angiogenesis | [122] |
| miR-491-3p | ABCB1, Sp3 | Drug resistance | [123] |
| miR-495 | IGF1R | Proliferation, invasion | [124] |
| miR-497 | VEGFA, AEG-1 | Angiogenesis, metastasis | [125] |
| miR-503 | No mentioned | Drug resistance | [126] |
| miR-506 | ROCK1 | Proliferation, apoptosis | [127] |
| miR-520f | TM4SF1 | Proliferation, invasion, migration | [128] |
| miR-539 | FSCN1 | Migration, invasion, drug resistance | [129, 130] |
| miR-542-3p | FZD7, Survivin | Proliferation | [131, 132] |
| miR-613 | YWHAZ | Proliferation, invasion | [133] |
| miR-634 | Rab1A, DHX33 | Cell growth, metastasis | [134] |
| miR-638 | VEGF, SOX2 | Angiogenesis, invasion, EMT | [135, 136] |
| miR-663a | HMGA2 | Proliferation, invasion | [137] |
| miR-708 | SMDAD3 | Proliferation, migration, invasion | [138] |
| miR-874 | DOR | Proliferation, metastasis | [139] |
| miR-874-3p | PIN1 | Proliferation, apoptosis | [140] |
| miR-876-5p | BCORL1 | Migration, invasion, EMT | [141] |
| miR-940 | CXCR2 | Migration, invasion, prognostic marker | [142] |
| miR-1207-5p | FASN | Cell growth, invasion | [143] |
| miR-1271-5p | FOXK2 | Cell growth, prognosis marker | [144] |
| miR-1299 | CDK6 | Proliferation | [145] |
| miR-1301 | BCL9, β-catenin, VEGFA | Migration, invasion, angiogenesis | [146] |
Recent studies have indicated that aberrant expressions of miRNAs were linked to HCC cells proliferation and apoptosis. Some miRNAs promoted cell proliferation and apoptosis of HCC, and the others were repressive. Therefore, these miRNAs can be as potential cancer inhibitors to control the development and progression of HCC by regulating cell proliferation and apoptosis.
Plenty of miRNAs were reported that they could mediate cell proliferation and apoptosis by controlling cell cycle in HCC. Overexpression of miR-1468 promoted cell cycle transition from G1 to S phase and apoptosis resistance [51]. Increased expression of miR-98 arrested HCC cell cycle in G0/G1 phase to repress cell proliferation via targeting enhancer of zeste homolog 2 (EZH2) [66]. Overexpression of miR-195 induced G1 phase cell cycle arrest and promoted apoptosis by directly targeting Wnt3 in HCC [91]. MiR-506 was reported to induce HCC cell cycle G1/S phase arrest and apoptosis [127]. MiR-1299 overexpression inhibited HCC cell cycle from G0/G1 phase entering into S phase, and its target cyclin dependent kinase 6 (CDK6) was the key regulator in the G0/G1 phase arrest [145].
Some aberrant expression of miRNAs could promote HCC cell proliferation and apoptosis by binding to their target genes. Inhibition of miR-25 expression was showed via PTEN/PI3K/Akt/Bad signaling pathway to enhance HCC cells apoptosis caused by the tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) [23]. MiR-107 was observed to be upregulated in HCC. Zhang JJ et al. reported that overexpression of miR-107 contributed to HCC cells proliferation via targeting Axin2 [27]. However, Wang Y et al. got that repressing miR-107 by targeting high mobility group A2 (HMGA2) could increase HCC cells proliferation [28]. MiR-155-5p was found to elevate HCC cells proliferation ability but inhibit apoptosis [31]. High expression of miR-203a-3p.1 could improve HCC cell proliferation by targeting interleukin-24 (IL-24) in HCC [35].
In addition, other miRNAs, which abnormally express, were proved to inhibit cell proliferation and apoptosis in HCC. MiR-96-5p was observed to inhibit apoptosis by targeting caspase-9 [26]. MiR-122 appeared abundant and downregulated in HCC cells. Xu Q et al. reported that overexpression of miR-122 repressed proliferation but induced apoptosis by targeting pyruvate kinase muscle 2 (PKM2) in HCC [72]. Another report found that miR-122 could downregulate the expression of oncogenic distal-less 4 (DLX4), knockdown the expression of this oncogene would inhibit HCC cells proliferation [73]. Overexpression of miR-137 was reported to reduce HepG2 cells proliferation by targeting EZH2 [76]. MiR-217 overexpression was revealed to inhibit cells apoptosis by targeting metadherin (MTDH) in HCC [105]. MiR-337 and miR-370 overexpression were also found to inhibit cell proliferation and promote apoptosis in HCC by HMGA2 and PIM1 [110] [118]. Decreased levels of miR-377 could suppress HCC cell apoptosis through inhibiting Bcl-xL expression [120].
The above reports have shown that miRNAs serve important roles in proliferation and apoptosis of liver cancer. In addition, it was indicated that many miRNAs participate in the development and progression of HCC by mediating proliferation and apoptosis.
MiRNAs and metastasis of HCC
Invasion and metastasis are the essential characteristics of the tumor cells. Metastasis is one of the most dominative causes of cancer death [152]. And 90% of cancer deaths are because of metastasis [153]. Tumor metastasis is a very complex process; it usually undergoes these major procedures: (1) local migration and infiltration, (2) vascular invasion, (3) survival in the circulating blood, (4) homing and implantation of metastatic organs in the distant place, (5) substantial infiltration, (6) adaptation to new environment, (7) secondary tumor growth [154]. EMT is a biological process that epithelial cells transform into mesenchymal cells by a specific procedure [155]. EMT participates in cancer metastasis through empowering tumor cells with migratory and invasive biological properties [156].
The latest researches have demonstrated that miRNAs can regulate HCC cells by promoting or suppressing HCC cells invasion, EMT and metastasis. How to prevent tumor metastasis has become one of the most important problems in the treatment of HCC. The discoveries of miRNAs may provide us with choices of anti-metastatic therapies.
These miRNAs dysregulated expression would contribute to HCC metastasis. MiR-25 overexpression could facilitate EMT formation by inhibiting Rho GDP dissociation inhibitor alpha (RhoGDI1) in HCC [22]. Overexpression of miR-135a promoted HCC cells migration and invasion by targeting forkhead box O1 (FOXO1) [30]. High expression of miR-203a-3p.1 could improve HCC migration and invasion by targeting IL-24 in HCC [35]. Upregulated miR-892a [48] and miR-1246 [49] expression were observed to enhance HCC cells migration and invasion. Downregulation miR-30e was showed to heighten metastasis and EMT of HCC cells by enhancing MTA1 [60]. Additionally, loss levels of miR-345 [114] and miR-638 [136] would heighten invasion and EMT of HCC cells.
Above miRNAs aberrant expression could motivate cell invasion, EMT and metastasis in HCC. Of course, some were the opposite. Downregulation of miR-197 was identified to inhibit HCC cells migration and invasion by targeting KAI1/CD82 [34]. Overexpression of miR-214-5p could inhibit the migration and invasion of HCC cells; besides, miR-214-5p could also suppress EMT [38]. MiR-212 overexpression was observed to inhibit the migration of HCC cells by targeting forkhead box M1 (FOXM1) and suppress the Wnt/β-Catenin signaling pathway [104]. MiR-495 and miR-613 overexpression were showed to inhibit cell proliferation and invasion in HCC by targeting IGF1R and YWHAZ [124] [133]. Upregulation of miR-122 expression in HCC repressed cell proliferation, invasion and EMT by targeting Snail1 and Snail2 [71]. Overexpression of miR-187-3p [89], miR-199b-5p [98] and miR-1301 [146] in HCC were also reported to inhibit EMT and metastasis. Besides, some miRNAs overexpression could repress invasion, migration and metastasis in HCC, for instance, miR-137 [76], miR-146a [85], miR-186 [88], miR-199 [94], miR-365 [116], miR-370 [118], miR-520f [128], miR-634 [134], miR-1207-5p [143], and so on.
Thus, miRNAs have been demonstrated to regulate metastasis of HCC. Absolutely, these miRNAs might be used to treat metastasis in HCC.
MiRNAs and angiogenesis of HCC
Abundant angiogenesis provides the necessary nutrition for tumor growth and metastasis, thus, it is essential for tumor growth and metastasis in solid tumor [157]. As one of the common solid tumors, HCC usually has affluent and deformed blood vessel tissue [158]. In the process of angiogenesis, vascular endothelial growth factor (VEGF), a highly conserved homodimeric glycoproteina, is identified as one of the most effective cytokines [159]. VEGF is a superfamily with seven subtypes, for example, VEGF-A, VEGF-B, VEGF-C, and so on. VEGF receptors have three types, VEGFR1, VEGFR2 and VEGFR3. VEGF family members combine with their receptors VEGFRs to induce tumor angiogenesis [160]. High expression of VEGF in tumor tissue or circulation blood frequently implies tumor may be invasion and metastasis [161].
Great deals of miRNAs were reported to regulate angiogenesis in HCC by VEGF. Overexpression of miR-146a was showed to repress HCC angiogenesis and tumor metastasis by downregulating VEGF [85]. MiR-199a-3p was proved to repress angiogenesis by directly decreasing VEGF secretion and suppressing expression of its receptors VEGFR1 and VEGFR2 on HCC cells [95]. MiR-451 could suppress VEGF production and block VEGFR2 pathway to reduce angiogenesis [122]. Overexpression of miR-638 was reported to suppress angiogenesis and tumor growth of HCC cells by inhibiting VEGF in HCC [135]. MiR-1301 overexpression was found to inhibit HCC angiogenesis by downregulating VEGFA, BCL9, and β-catenin [146].
On the contrary, some miRNAs could enhance angiogenesis by VEGF. Suppression miR-338-3p could upregulate VEGF expression to promote angiogenesis in HCC [112]. Furthermore, downregulating miR-497 promoted angiogenesis and metastasis by directly inhibiting VEGFA [125].
In consequence, miRNAs were proved the vital regulators in the process of HCC angiogenesis. What's more, miRNAs could act as inhibitors of tumor angiogenesis.
MiRNAs and drug resistance of HCC
Chemotherapy is currently one of the most commonly used treatment methods, when most patients with HCC are diagnosed at advanced stages [162]. Large numbers of trials that tested the efficacy of various drugs have manifested that HCC has low sensitivity to chemotherapy [163]. And several chemotherapies fail due to the intrinsic or acquired drug resistance [164]. Thus, how to reverse drug resistance and improve the effectiveness of chemotherapy are crucial problems to be solved urgently. Many reports have showed that miRNAs could act as regulators to promote or reverse drug resistance in HCC, indicating miRNAs might have the promising therapeutic potential for drug resistance.
Sorafenib is as known the first-line drug for advanced HCC, but its curative effect is limited due to acquired resistance, which may be the primary factor [165]. MiRNAs could reverse this effect. MiR-7 was proved to overcome sorafenib resistance by suppressing its target TYRO3 via PI3-Kinase/AKT pathway [55]. Overexpression of miR-216a/217 activated TGF-β pathway to induce sorafenib resistance, but interdicting TGF-β pathway would reverse this resistance in HCC [39]. MiR-367-3p increased sorafenib efficacy to suppress HCC metastasis through changing the MDM2/AR/FKBP5/PHLPP/ (pAKT and pERK) signals [117]. Another report has showed that sorafenib significantly reduced miR-142-3p levels by acting on the transcription factor PU.1; however, miR142-3p upregulation could sensitize HCC cells to sorafenib through targeting autophagy-related 5 (ATG5) and autophagy-related 16-like 1 (ATG16L1) to reduce sorafenib-induced autophagy, enhance sorafenib-induced apoptosis and inhibit cell growth [82].
For the drug resistance induced by other chemotherapeutic drugs, miRNAs also can promote or reverse drug resistance in HCC. Upregulating miR-182 was observed to increase cisplatin resistance in HCC treatment by regulating tumor protein 53-induced nuclear protein1 (TP53INP1) [33]. Inhibition of miR-33a-5p expression could also reduce cisplatin sensitivity and increased its drug resistance in HCC [64]. MiR-7/21/107 was enhanced by HBV X protein to promote HCC cells drug resistance by directly suppressing its target maspin expression [56]. MiR-31 and its target gene NDRG3 made HCC cells sensitize to chemotherapeutic drug Adriamycin [61]. MiR-375 was combined with hollow mesoporous silica nanoparticles (HMSN) to overcome doxorubicin hydrochloride resistance in HCC [119]. Additionally, miR-539 overexpression was reported to increase sensitivity to antagonize arsenic trioxide resistance in HCC [130].
Therefore, miRNAs were involved in drug resistance of HCC. Furthermore, miRNAs could prove a new therapeutic strategy for how to improve the effectiveness of chemotherapy when treating HCC.
MiRNAs and autophagy of HCC
Autophagy has been reported for many years ago, but it has recently gained more attention, especially the Nobel Prize in Physiology or Medicine awarding to the great discovery of autophagy in 2016 makes it a popular topic again. Autophagy, a self-digestive catabolism process [166], depends on lysosomes to degrade and recycle proteins or cell organelles [167]. Autophagy can regulate cell survival, differentiation, senescence, death and many other biological processes [168]. It has been proved that autophagy has a dual regulation role in HCC occurrence and suppression [169].
MiRNAs might participate in the process of HCC development and progression through autophagy. MiR-181a was reported to repress autophagy in HCC by targeting pro-autophagic protein Atg5, leading to reducing apoptosis of HCC cells and accelerate hepatoma growth [32]. MiR-7 was confirmed to induce HCC cells autophagy by targeting mammalian target of rapamycin (mTOR), and inhibition of autophagy heightened the antitumor activity of miR-7 to repress HCC cells proliferation [54]. MiR-26 could improve HCC cells sensibility to chemotherapy and facilitated apoptosis of HCC cells through inhibiting autophagy initiator ULK1 [57]. MiR-101 was found to enhance cisplatin-induced apoptosis through repressing autophgy in HCC [68].
Thus, miRNAs participate in the process of HCC tumorigenesis and development through autophagy.
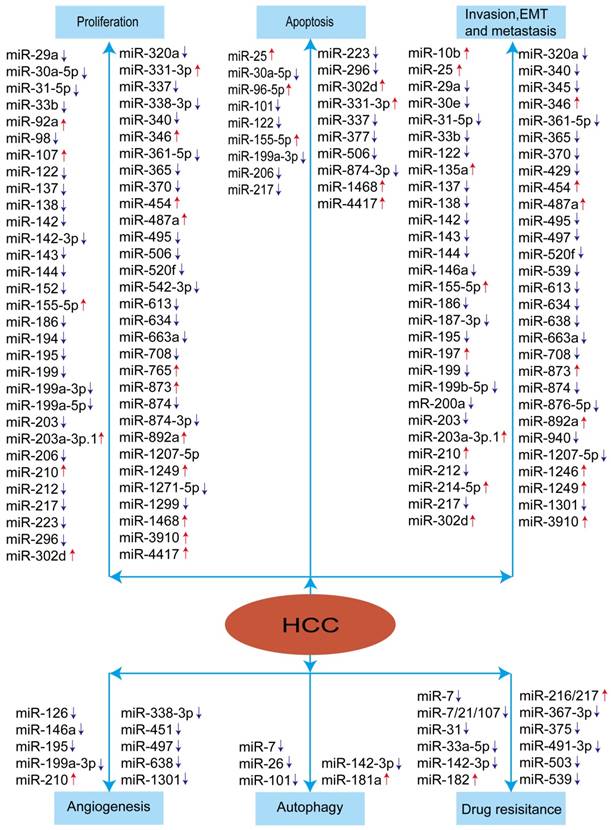
To sum up, miRNAs appear to play crucial roles in modulating HCC development and progression. The aberrant expression of miRNAs in HCC was summarized in the Figure 2. These studies indicated that miRNAs have the promising therapeutic potential for HCC treatment.
Exosomal miRNAs and HCC
Exosomes, one type in extracellular vesicles (EVs), are small vesicles with a size range of 40-150 nm and a lipid bilayer membrane [170]. Exosomes, which now considered as an additional mechanism for intercellular communication [171], are generated inside multivesicular endosomes or multivesicular bodies (MVBs) [172]. Exosomes exist in all body fluids, such as serum, urine, and saliva [173]. Exosomes have been shown to act as shuttles between cells including RNA, proteins, miRNAs, long noncoding RNAs (lncRNAs), or DNA fragments [174-177]. Tumor-derived exosomes are recognized as a critical determinant of the tumor progression [178]. Studies have demonstrated the mechanism of HCC-derived exosome-mediated miRNA transfer is important in the growth and progression of HCC [179]. The studies of exosomal miRNAs in HCC in recent years were summarized in the Table 3 [180-194].
Summary of miRNAs in the development and progression of HCC. Red arrow means: increased expression of miRNA, blue arrow means: decreased expression of miRNA.
Exosomal miRNAs were involved in proliferation, migration, metastasis, drug resistance in HCC. Exosomal miR-9-3p, lower level in HCC patients, could reduce HCC cell viability and proliferation, and additionally reduced ERK1/2 expression by targeting fibroblast growth factor 5 (HBGF-5) [180]. Exosomal miR-32-5p was testified to activate the PI3K/Akt pathway, and induce multidrug resistance by modulating angiogenesis and EMT in HCC [183]. Exosomal miR-103 was proved to increase vascular permeability and promote metastasis by targeting junction proteins [184]. Zhang Z et al. found that the expression of exosomal miR-320a in cancer-associated fibroblasts (CAFs) was lower than paracancer fibroblasts (PAFs), leading to the cancer cells towards a more malignant phenotype. Furthermore, they revealed that miR-320a could suppress HCC cell proliferation, migration and metastasis by directly targeting PBX3 [186]. Tumor-derived exosomal miR-1247-3p was observed to convert fibroblasts to cancer-associated fibroblasts (CAFs) via downregulating B4GALT3, to activate β1-integrin-NF-κB signaling pathway to promote lung metastasis of liver cancer [190].
Exosomal miRNAs in HCC
| Exosomal miRNA | Targets | Mechanisms | References |
|---|---|---|---|
| miR-9-3p | HBGF-5 | Proliferation | [180] |
| miR-21 | No mentioned | Diagnostic marker | [181] |
| miR-26a | No mentioned | Proliferation, migration | [182] |
| miR-32-5p | PTEN | Drug resistance | [183] |
| miR-103 | VE-Cad, p120 ZO-1 | Metastasis | [184] |
| miR-122 | ADAM10, IGF1R, CCNG1 | Drug resistance | [185] |
| miR-320a | PBX3 | Proliferation, migration, metastasis | [186] |
| miR-335-5p | No mentioned | Proliferation, invasion | [187] |
| miR-638 | No mentioned | Prognosis marker | [188] |
| miR-718 | HOXB8 | Proliferation, prognostic marker | [189] |
| miR-1247-3p | B4GALT3 | Metastasis | [190] |
| miR-122, miR-148a, miR-1246 | No mentioned | Diagnostic marker | [191] |
| miR-18a, miR-221, miR-222, miR-224, miR-101, miR-106b, miR-122, miR-195 | No mentioned | Diagnostic marker | [192] |
| miR-10b, miR-21, miR-122, miR-200a | No mentioned | Diagnostic marker | [193] |
| miR-519d, miR-21, miR-221, miR-1228 | No mentioned | Diagnostic marker | [194] |
Therefore, exosomes can transfer miRNAs between cells, and these miRNAs play important roles in proliferation, invasion, metastasis, and drug resistance in HCC by regulating gene expression in the target cells.
Acting as diagnostic and prediction markers in HCC
When HCC are diagnosed, many patients have already been advanced stage. It would have far-reaching influence on the prevention and treatment of HCC if the cancer could be early diagnosed and detected. It is well recognized that alpha fetoprotein (AFP) is the most common used hematology diagnosis marker of HCC in the clinical. But its false negative rate may be 40% with early stage HCC [195]. Moreover, some non-tumor diseases, such as hepatitis and cirrhosis, these patients' serum APF levels may also elevate [196] .Therefore, it is necessary to seek for some new markers to diagnose and predict HCC. MiRNAs may have the potential functions according to the above mechanisms.
Numerous researches have supported that miRNAs could act as diagnostic and prediction markers in HCC. MiRNAs could be used to diagnose and distinguish HCC. For example, miR-101 levels in the serum were found to be significantly downregulated in the HBV-related HCC patients compared with the HBV-related liver cirrhosis patients, chronic hepatitis B patients and healthy controls, indicating that miR-101 could severe as a potential hematological marker of to diagnose and distinguish HBV-related HCC [69]. MiRNAs could also diagnose tumor size and TNM stages of HCC. High miR‑32 expression was observed that large tumor size (≥ 5cm) had significantly decreased [24]. High expression of miR-1246 and its target gene CADM1 low expression were correlated with stage 1 of TNM stages in HCC [49]. Low expression of miR-296 in HCC patients might have large tumor size and advanced TNM stage [108]. Low expression of miR‑137 was significantly closely related with lymph node metastasis, vein invasion and advanced clinical stage in HCC [76].
Besides, miRNAs were reported to be as useful prognosis markers as well. High expression of miR-92a [25], miR-221 [40], miR-487a [45] and miR-1468 [51] might indicate poor prognosis in HCC. Low expression of miR-33a [63], miR-122 [72], miR-137 [76], miR-194 [90] and miR-940 [142] in HCC patients were observed to have unfavorable prognosis. High levels of miR-7/21/107 and low expression of maspin implied poor survival of HBV-related HCC [56]. Low expression of miR-138 combined with high expression of its target cyclin D3 showed worse clinical prognosis in HCC [77]. High expression of forkhead box K2 (FOXK2) protein, the target of miR-1271-5p, had poor overall survival (OS) and disease-free survival (DFS) of HCC patients [144].
Exosomal miRNAs have been as novel biomarkers for HCC diagnoses and prognosis in clinical in recent years. For example, high expression of exosomal miR-32-5p and low expression of its target PTEN were positively associated with poor prognosis [183]. HCC patients with lower levels of serum exosomal miR-638 had poor overall survival than those with higher levels of exosomal miR-638 in serum [188]. The expression level of serum exosomal miR-21 was significantly higher in patients with HCC than those with chronic hepatitis B (CHB) or healthy volunteers. Besides, high level of miR-21 expression correlated with cirrhosis and advanced tumor stage [181]. MiR-122, miR-148a, and miR-1246 were significantly elevated in serum exosomes from HCC patients compared to liver cirrhosis (LC) and normal control (NC) individuals [191].
Taken together, all of the above researches suggested that these miRNAs, including exosomal miRNAs, could be valuable of diagnostic and prediction markers in HCC.
Conclusions and future directions
Great progress has been made in the study of miRNAs in HCC. MiRNAs are pivotal participants and regulators in the development and progression of HCC. Proliferation, apoptosis, invasion, metastasis, EMT, angiogenesis, drug resistance and autophagy of miRNAs may be the primary regulatory mechanisms in HCC. Exosomal miRNAs have been focused on in recent years, and the researches are progressing rapidly. Recent studies have shown that exosomes can transfer miRNAs between cells in proliferation, invasion, metastasis, and drug resistance of HCC. In addition, exosomal miRNAs could be as biomarkers for HCC diagnoses and prognosis. The above studies indicated miRNAs could be used valid therapeutic targets and acted as valuable early diagnostic and prediction markers in HCC. Understanding the regulatory mechanisms of miRNAs in the HCC development and progression will help us to develop more effective new therapies and molecular therapeutic drugs.
However, there are also some problems in the studies of miRNAs. Lots of studies only stay in the experimental stage and do not really be used into the clinic. Secondly, the security and reliability of miRNAs acting as HCC early diagnosis and treatment markers also need to further research. In addition, Exosomal miRNAs are mostly concentrated in the observation of the content of miRNAs in serum exosomes, but their specific mechanisms of HCC are not fully understood. And the lacks of sensitive preparatory and analytical technologies for exosomes are also big challenges to clinical translation [197].
Therefore, the future studies should pay more attention to make the acquired achievement of miRNAs in HCC translate into clinical application, for instance, develop available miRNA inhibitors for clinical. Besides, the research in security and reliability of miRNAs for HCC early diagnosis and treatment should also be more concerned. For the research of exosomal miRNAs in HCC, the mechanisms of exosome-mediated miRNAs transfer should be focused on in the future studies. Meanwhile, the analytical technologies also need to be improved.
Acknowledgements
This study was supported by Natural science foundation of China (Grants 81472124 and 81774291), “Chen Guang” project supported by Shanghai Municipal Education Commission and Shanghai Education Development Foundation (Grant 17CG 43, to Lifang Ma) and Talent introduction project of Shanghai Municipal Hospital of Traditional Chinese Medicine (Grant 20160501, to Lifang Ma). Innovation project of Shanghai University of Traditional Chinese Medicine (Grant JXDXSCXJH15, to Yuquan Tao).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-86
2. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108
3. Tunissiolli NM, Castanhole-Nunes MMU, Biselli-Chicote PM, Pavarino EC, da Silva RF, da Silva RC. et al. Hepatocellular carcinoma: A comprehensive review of biomarkers, clinical aspects, and therapy. Asian Pac J Cancer Prev. 2017;18:863-72
4. Xia F, Wu LL, Lau WY, Huan HB, Wen XD, Ma KS. et al. Adjuvant sorafenib after heptectomy for Barcelona Clinic Liver Cancer-stage C hepatocellular carcinoma patients. World J Gastroenterol. 2016;22:5384-92
5. Gosalia AJ, Martin P, Jones PD. Advances and future directions in the treatment of hepatocellular carcinoma. Gastroenterol Hepatol (N Y). 2017;13:398-410
6. Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF. et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378-90
7. Bruix J, Qin S, Merle P, Granito A, Huang YH, Bodoky G. et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;389:56-66
8. International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931-45
9. Weng M, Wu D, Yang C, Peng H, Wang G, Wang T. et al. Noncoding RNAs in the development, diagnosis, and prognosis of colorectal cancer. Transl Res. 2017;181:108-20
10. Rupaimoole R, Calin GA, Lopez-Berestein G, Sood AK. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov. 2016;6:235-46
11. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-33
12. Shukla GC, Singh J, Barik S. MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011;3:83-92
13. Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH. et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051-60
14. Li J, Tian H, Yang J, Gong Z. Long noncoding RNAs regulate cell growth, proliferation, and apoptosis. DNA Cell Biol. 2016;35:459-70
15. Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95-8
16. Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J. et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415-9
17. Kim Y, Kim VN. MicroRNA factory: RISC assembly from precursor microRNAs. Mol Cell. 2012;46:384-6
18. Kanellopoulou C, Monticelli S. A role for microRNAs in the development of the immune system and in the pathogenesis of cancer. Semin Cancer Biol. 2008;18:79-88
19. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203-22
20. Zhu Q, Gong L, Wang J, Tu Q, Yao L, Zhang JR. et al. miR-10b exerts oncogenic activity in human hepatocellular carcinoma cells by targeting expression of CUB and sushi multiple domains 1 (CSMD1). BMC Cancer. 2016;16:806
21. Koenig AB, Barajas JM, Guerrero MJ, Ghoshal K. A Comprehensive Analysis of Argonaute-CLIP Data Identifies Novel, Conserved and Species-Specific Targets of miR-21 in Human Liver and Hepatocellular Carcinoma. Int J Mol Sci. 2018;19:851
22. Wang C, Wang X, Su Z, Fei H, Liu X, Pan Q. miR-25 promotes hepatocellular carcinoma cell growth, migration and invasion by inhibiting RhoGDI1. Oncotarget. 2015;6:36231-44
23. Feng X, Jiang J, Shi S, Xie H, Zhou L, Zheng S. Knockdown of miR-25 increases the sensitivity of liver cancer stem cells to TRAIL-induced apoptosis via PTEN/PI3K/Akt/Bad signaling pathway. Int J Oncol. 2016;49:2600-10
24. Yang H, Li Y, Zhong X, Luo P, Luo P, Sun R. et al. Upregulation of microRNA-32 is associated with tumorigenesis and poor prognosis in patients with hepatocellular carcinoma. Oncol Lett. 2018;15:4097-104
25. Yang W, Dou C, Wang Y, Jia Y, Li C, Zheng X. et al. MicroRNA-92a contributes to tumor growth of human hepatocellular carcinoma by targeting FBXW7. Oncol Rep. 2015;34:2576-84
26. Iwai N, Yasui K, Tomie A, Gen Y, Terasaki K, Kitaichi T. et al. Oncogenic miR-96-5p inhibits apoptosis by targeting the caspase-9 gene in hepatocellular carcinoma. Int J Oncol. 2018 [Epub ahead of print]
27. Zhang JJ, Wang CY, Hua L, Yao KH, Chen JT, Hu JH. miR-107 promotes hepatocellular carcinoma cell proliferation by targeting Axin2. Int J Clin Exp Pathol. 2015;8:5168-74
28. Wang Y, Chen F, Zhao M, Yang Z, Zhang S, Ye LH. et al. MiR-107 suppresses proliferation of hepatoma cells through targeting HMGA2 mRNA 3' UTR. Biochem Biophys Res Commun. 2016;480:455-60
29. Su SG, Yang M, Zhang MF, Peng QZ, Li MY, Liu LP. et al. miR-107-mediated decrease of HMGCS2 indicates poor outcomes and promotes cell migration in hepatocellular carcinoma. Int J Biochem Cell Biol. 2017;91:53-9
30. Zeng YB, Liang XH, Zhang GX, Jiang N, Zhang T, Huang JY. et al. miRNA-135a promotes hepatocellular carcinoma cell migration and invasion by targeting forkhead box O1. Cancer Cell Int. 2016;16:63
31. Fu X, Wen H, Jing L, Yang Y, Wang W, Liang X. et al. MicroRNA-155-5p promotes hepatocellular carcinoma progression by suppressing PTEN through the PI3K/Akt pathway. Cancer Sci. 2017;108:620-31
32. Yang J, He Y, Zhai N, Ding S, Li JP, Peng Z. MicroRNA-181a inhibits autophagy by targeting Atg5 in hepatocellular carcinoma. Front Biosci (Landmark Ed). 2018;23:388-96
33. Qin J, Luo M, Qian H, Chen W. Upregulated miR-182 increases drug resistance in cisplatin-treated HCC cell by regulating TP53INP1. Gene. 2014;538:342-7
34. Dai W, Wang C, Wang F, Wang Y, Shen M, Chen K. et al. Anti-miR-197 inhibits migration in HCC cells by targeting KAI 1/CD82. Biochem Biophys Res Commun. 2014;446:541-8
35. Huo W, Du M, Pan X, Zhu X, Gao Y, Li Z. miR-203a-3p.1 targets IL-24 to modulate hepatocellular carcinoma cell growth and metastasis. FEBS Open Bio. 2017;7:1085-91
36. Yang Y, Zhang J, Xia T, Li G, Tian T, Wang M. et al. MicroRNA-210 promotes cancer angiogenesis by targeting fibroblast growth factor receptor-like 1 in hepatocellular carcinoma. Oncol Rep. 2016;36:2553-62
37. Tan W, Lim SG, Tan TM. Up-regulation of microRNA-210 inhibits proliferation of hepatocellular carcinoma cells by targeting YES1. World J Gastroenterol. 2015;21:13030-41
38. Li H, Wang H, Ren Z. MicroRNA-214-5p Inhibits the Invasion and Migration of Hepatocellular Carcinoma Cells by Targeting Wiskott-Aldrich Syndrome Like. Cell Physiol Biochem. 2018;46:757-64
39. Xia H, Ooi LL, Hui KM. MicroRNA-216a/217-induced epithelial-mesenchymal transition targets PTEN and SMAD7 to promote drug resistance and recurrence of liver cancer. Hepatology. 2013;58:629-41
40. Chen F, Li XF, Fu DS, Huang JG, Yang SE. Clinical potential of miRNA-221 as a novel prognostic biomarker for hepatocellular carcinoma. Cancer Biomark. 2017;18:209-14
41. Chen YL, Xu QP, Guo F, Guan WH. MicroRNA-302d downregulates TGFBR2 expression and promotes hepatocellular carcinoma growth and invasion. Exp Ther Med. 2017;13:681-7
42. Cao Y, Chen J, Wang D, Peng H, Tian X, Xiong D. et al. Upregulated in Hepatitis B virus-associated hepatocellular carcinoma cells, miR-331-3p promotes proliferation of hepatocellular carcinoma cells by targeting ING5. Oncotarget. 2015;6:38093-106
43. Yu Q, Yang X, Duan W, Li C, Luo Y, Lu S. miRNA-346 promotes proliferation, migration and invasion in liver cancer. Oncol Lett. 2017;14:3255-60
44. Yu L, Gong X, Sun L, Yao H, Lu B, Zhu L. miR-454 functions as an oncogene by inhibiting CHD5 in hepatocellular carcinoma. Oncotarget. 2015;6:39225-34
45. Chang RM, Xiao S, Lei X, Yang H, Fang F, Yang LY. miRNA-487a promotes proliferation and metastasis in hepatocellular carcinoma carcinoma. Clin Cancer Res. 2017;23:2593-604
46. Xie BH, He X, Hua RX, Zhang B, Tan GS, Xiong SQ. et al. Mir-765 promotes cell proliferation by downregulating INPP4B expression in human hepatocellular carcinoma. Cancer Biomark. 2016;16:405-13
47. Han G, Zhang L, Ni X, Chen Z, Pan X, Zhu Q. et al. MicroRNA-873 Promotes Cell Proliferation, Migration, and Invasion by Directly Targeting TSLC1 in Hepatocellular Carcinoma. Cell Physiol Biochem. 2018;46:2261-70
48. Jia B, Tan L, Jin Z, Jiao Y, Fu Y, Liu Y. MiR-892a promotes hepatocellular carcinoma cells proliferation and invasion through targeting CD226. J Cell Biochem. 2017;118:1489-96
49. Sun Z, Meng C, Wang S, Zhou N, Guan M, Bai CM. et al. MicroRNA-1246 enhances migration and invasion through CADM1 in hepatocellular carcinoma. BMC Cancer. 2014;14:616
50. Ye Y, Wei Y, Xu Y, Li Y, Wang R, Chen J. et al. Induced MiR-1249 expression by aberrant activation of Hedegehog signaling pathway in hepatocellular carcinoma. Exp Cell Res. 2017;355:9-17
51. Liu Z, Wang Y, Dou C, Sun L, Li Q, Wang L. et al. MicroRNA-1468 promotes tumor progression by activating PPAR-gamma-mediated AKT signaling in human hepatocellular carcinoma. J Exp Clin Cancer Res. 2018;37:49
52. Cheng L, Wang H, Han S. MiR-3910 promotes the growth and migration of cancer cells in the progression of hepatocellular carcinoma. Dig Dis Sci. 2017;62:2812-20
53. Song L, Zhang W, Chang Z, Pan Y, Zong H, Fan Q. et al. miR-4417 targets tripartite motif-containing 35 (TRIM35) and regulates pyruvate kinase muscle 2 (PKM2) phosphorylation to promote proliferation and suppress apoptosis in hepatocellular carcinoma cells. Med Sci Monit. 2017;23:1741-50
54. Wang Y, Wang Q, Song J. Inhibition of autophagy potentiates the proliferation inhibition activity of microRNA-7 in human hepatocellular carcinoma cells. Oncol Lett. 2017;14:3566-72
55. Kabir TD, Ganda C, Brown RM, Beveridge DJ, Richardson KL, Chaturvedi V. et al. A microRNA-7/growth arrest specific 6/TYRO3 axis regulates the growth and invasiveness of sorafenib-resistant cells in human hepatocellular carcinoma. Hepatology. 2018;67:216-31
56. Chen WS, Yen CJ, Chen YJ, Chen JY, Wang LY, Chiu SJ. et al. miRNA-7/21/107 contribute to HBx-induced hepatocellular carcinoma progression through suppression of maspin. Oncotarget. 2015;6:25962-74
57. Jin F, Wang Y, Li M, Zhu Y, Liang H, Wang C. et al. MiR-26 enhances chemosensitivity and promotes apoptosis of hepatocellular carcinoma cells through inhibiting autophagy. Cell Death Dis. 2017;8:e2540
58. Mahati S, Xiao L, Yang Y, Mao R, Bao Y. miR-29a suppresses growth and migration of hepatocellular carcinoma by regulating CLDN1. Biochem Biophys Res Commun. 2017;486:732-7
59. He R, Yang L, Lin X, Chen X, Lin X, Wei F. et al. MiR-30a-5p suppresses cell growth and enhances apoptosis of hepatocellular carcinoma cells via targeting AEG-1. Int J Clin Exp Pathol. 2015;8:15632-41
60. Deng L, Tang J, Yang H, Chen C, Lu S, Jiang R. et al. MTA1 modulated by miR-30e contributes to epithelial-to-mesenchymal transition in hepatocellular carcinoma through an ErbB2-dependent pathway. Oncogene. 2017;36:3976-85
61. Du Z, Niu S, Xu X, Xu Q. MicroRNA31-NDRG3 regulation axes are essential for hepatocellular carcinoma survival and drug resistance. Cancer Biomark. 2017;19:221-30
62. Zhao G, Han C, Zhang Z, Wang L, Xu J. Increased expression of microRNA-31-5p inhibits cell proliferation, migration, and invasion via regulating Sp1 transcription factor in HepG2 hepatocellular carcinoma cell line. Biochemical and Biophysical Research Communications. 2017;490:371-7
63. Xie RT, Cong XL, Zhong XM, Luo P, Yang HQ, Lu GX. et al. MicroRNA-33a downregulation is associated with tumorigenesis and poor prognosis in patients with hepatocellular carcinoma. Oncol Lett. 2018;15:4571-7
64. Meng W, Tai Y, Zhao H, Fu B, Zhang T, Liu W. et al. Downregulation of miR-33a-5p in hepatocellular carcinoma: A possible mechanism for chemotherapy resistance. Med Sci Monit. 2017;23:1295-304
65. Tian Q, Xiao Y, Wu Y, Liu Y, Song Z, Gao W. et al. MicroRNA-33b suppresses the proliferation and metastasis of hepatocellular carcinoma cells through the inhibition of Sal-like protein 4 expression. Int J Mol Med. 2016;38:1587-95
66. Zhang JJ, Chen JT, Hua L, Yao KH, Wang CY. miR-98 inhibits hepatocellular carcinoma cell proliferation via targeting EZH2 and suppressing Wnt/β-catenin signaling pathway. Biomed Pharmacother. 2017;85:472-8
67. He H, Tian W, Chen H, Deng Y. MicroRNA-101 sensitizes hepatocellular carcinoma cells to doxorubicin-induced apoptosis via targeting Mcl-1. Mol Med Rep. 2016;13:1923-9
68. Xu Y, An Y, Wang Y, Zhang C, Zhang H, Huang C. et al. miR-101 inhibits autophagy and enhances cisplatin-induced apoptosis in hepatocellular carcinoma cells. Oncol Rep. 2013;29:2019-24
69. Xie Y, Yao Q, Butt AM, Guo J, Tian Z, Bao X. et al. Expression profiling of serum microRNA-101 in HBV-associated chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. Cancer Biol Ther. 2014;15:1248-55
70. Ma YS, Wu TM, Lv ZW, Lu GX, Cong XL, Xie RT. et al. High expression of miR-105-1 positively correlates with clinical prognosis of hepatocellular carcinoma by targeting oncogene NCOA1. Oncotarget. 2017;8:11896-905
71. Jin Y, Wang J, Han J, Luo D, Sun Z. MiR-122 inhibits epithelial-mesenchymal transition in hepatocellular carcinoma by targeting Snail1 and Snail2 and suppressing WNT/beta-cadherin signaling pathway. Exp Cell Res. 2017;360:210-7
72. Xu Q, Zhang M, Zhang J, Pang L, Cai W, Liu X. MicroRNA-122 affects cell aggressiveness and apoptosis by targeting PKM2 in human hepatocellular carcinoma. Oncol Rep. 2015;34:2054-64
73. Xie XH, Xu XP, Sun CY, Yu ZJ. Regulation of the oncogenic function of distal-less 4 by microRNA-122 in hepatocellular carcinoma. Mol Med Rep. 2015;12:1375-80
74. He RQ, Yang X, Liang L, Chen G, Ma J. MicroRNA-124-3p expression and its prospective functional pathways in hepatocellular carcinoma: A quantitative polymerase chain reaction, gene expression omnibus and bioinformatics study. Oncol Lett. 2018;15:5517-32
75. Jing BQ, Ou Y, Zhao L, Xie Q, Zhang YX. Experimental study on the prevention of liver cancer angiogenesis via miR-126. Eur Rev Med Pharmacol Sci. 2017;21:5096-100
76. Cui S, Sun Y, Liu Y, Liu C, Wang J, Hao G. et al. MicroRNA137 has a suppressive role in liver cancer via targeting EZH2. Mol Med Rep. 2017;16:9494-502
77. Huang B, Li H, Huang L, Luo C, Zhang Y. Clinical significance of microRNA 138 and cyclin D3 in hepatocellular carcinoma. J Surg Res. 2015;193:718-23
78. Liu C, Zhu J, Liu F, Wang Y, Zhu M. MicroRNA-138 targets SP1 to inhibit the proliferation, migration and invasion of hepatocellular carcinoma cells. Oncol Lett. 2018;15:1279-86
79. Su F, Zhao J, Qin S, Wang R, Li Y, Wang Q. et al. Over-expression of Thrombospondin 4 correlates with loss of miR-142 and contributes to migration and vascular invasion of advanced hepatocellular carcinoma. Oncotarget. 2017;8:23277-88
80. Yu Q, Xiang L, Yin L, Liu X, Yang D, Zhou J. Loss-of-function of miR-142 by hypermethylation promotes TGF-beta-mediated tumour growth and metastasis in hepatocellular carcinoma. Cell Prolif. 2017;50:e12384
81. Hua S, Liu C, Liu L, Wu D. miR-142-3p inhibits aerobic glycolysis and cell proliferation in hepatocellular carcinoma via targeting LDHA. Biochem Biophys Res Commun. 2018;496:947-54
82. Zhang K, Chen J, Zhou H, Chen Y, Zhi Y, Zhang B. et al. PU.1/microRNA-142-3p targets ATG5/ATG16L1 to inactivate autophagy and sensitize hepatocellular carcinoma cells to sorafenib. Cell Death Dis. 2018;9:312
83. Liu X, Gong J, Xu B. miR-143 down-regulates TLR2 expression in hepatoma cells and inhibits hepatoma cell proliferation and invasion. Int J Clin Exp Pathol. 2015;8:12738-47
84. Bao HB, Li XG, Li HL, Xing HL, Xu BH, Zhang XF. et al. MicroRNA-144 inhibits hepatocellular carcinoma cell proliferation, invasion and migration by targeting ZFX. J Biosci. 2017;42:103-11
85. Zhang Z, Zhang Y, Sun XX, Ma X, Chen ZN. microRNA-146a inhibits cancer metastasis by downregulating VEGF through dual pathways in hepatocellular carcinoma. Mol Cancer. 2015;14:5
86. Zhou J, Zhang Y, Qi Y, Yu D, Shao Q, Liang J. MicroRNA-152 inhibits tumor cell growth by directly targeting RTKN in hepatocellular carcinoma. Oncol Rep. 2017;37:1227-34
87. Zhao Z, Hu Y, Shen X, Lao Y, Zhang L, Qiu X. et al. HBx represses RIZ1 expression by DNA methyltransferase 1 involvement in decreased miR-152 in hepatocellular carcinoma. Oncol Rep. 2017;37:2811-8
88. Ruan T, He X, Yu J, Hang Z. MicroRNA-186 targets Yes-associated protein 1 to inhibit Hippo signaling and tumorigenesis in hepatocellular carcinoma. Oncol Lett. 2016;11:2941-5
89. Dou C, Liu Z, Xu M, Jia Y, Wang Y, Li Q. et al. miR-187-3p inhibits the metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by targeting S100A4. Cancer Lett. 2016;381:380-90
90. Zhao Y, Li F, Zhang X, Liu A, Qi J, Cui H. et al. MicroRNA-194 acts as a prognostic marker and inhibits proliferation in hepatocellular carcinoma by targeting MAP4K4. Int J Clin Exp Pathol. 2015;8:12446-54
91. Yang Y, Li M, Chang S, Wang L, Song T, Gao L. et al. MicroRNA-195 acts as a tumor suppressor by directly targeting Wnt3a in HepG2 hepatocellular carcinoma cells. Mol Med Rep. 2014;10:2643-8
92. Zheng C, Li J, Wang Q, Liu W, Zhou J, Liu R. et al. MicroRNA-195 functions as a tumor suppressor by inhibiting CBX4 in hepatocellular carcinoma. Oncol Rep. 2015;33:1115-22
93. Wang M ZJ, Tong LL, Ma XF, Qiu XG. miR-195 is a key negative regulator of hepatocellular carcinoma metastasis by targeting FGF2 and VEGFA. Int J Clin Exp Pathol. 2015;8:14110-20
94. Zhang W, Qian S, Yang G, Zhu L, Zhou B, Wang J. et al. MicroRNA-199 suppresses cell proliferation, migration and invasion by downregulating RGS17 in hepatocellular carcinoma. Gene. 2018;659:22-8
95. Ghosh A, Dasgupta D, Ghosh A, Roychoudhury S, Kumar D, Gorain M. et al. MiRNA199a-3p suppresses tumor growth, migration, invasion and angiogenesis in hepatocellular carcinoma by targeting VEGFA, VEGFR1, VEGFR2, HGF and MMP2. Cell Death Dis. 2017;8:e2706
96. Ren K, Li T, Zhang W, Ren J, Li Z, Wu G. miR-199a-3p inhibits cell proliferation and induces apoptosis by targeting YAP1, suppressing Jagged1-Notch signaling in human hepatocellular carcinoma. J Biomed Sci. 2016;23:79
97. Huang GH, Shan H, Li D, Zhou B, Pang PF. MiR-199a-5p suppresses tumorigenesis by targeting clathrin heavy chain in hepatocellular carcinoma. Cell Biochem Funct. 2017;35:98-104
98. Zhou SJ, Liu FY, Zhang AH, Liang HF, Wang Y, Ma R. et al. MicroRNA-199b-5p attenuates TGF-beta1-induced epithelial-mesenchymal transition in hepatocellular carcinoma. Br J Cancer. 2017;117:233-44
99. Cui X, Li Z, Gao J, Gao PJ, Ni YB, Zhu JY. Elevated CXCL1 increases hepatocellular carcinoma aggressiveness and is inhibited by miRNA-200a. Oncotarget. 2016;7:65052-66
100. Wang J, Song W, Shen W, Yang X, Sun W, Qu S. et al. MicroRNA-200a suppresses cell invasion and migration by directly targeting GAB1 in hepatocellular carcinoma. Oncol Res. 2017;25:1-10
101. Zheng XB, Chen XB, Xu LL, Zhang M, Feng L, Yi PS. et al. miR-203 inhibits augmented proliferation and metastasis of hepatocellular carcinoma residual in the promoted regenerating liver. Cancer Sci. 2017;108:338-46
102. Wu H, Tao J, Li X, Zhang T, Zhao L, Wang Y. et al. MicroRNA-206 prevents the pathogenesis of hepatocellular carcinoma by modulating expression of met proto-oncogene and cyclin-dependent kinase 6 in mice. Hepatology. 2017;66:1952-67
103. Deng B, Qu L, Li J, Fang J, Yang S, Cao Z. et al. MiRNA-211 suppresses cell proliferation, migration and invasion by targeting SPARC in human hepatocellular carcinoma. Sci Rep. 2016;6:26679
104. Jia P, Wei G, Zhou C, Gao Q, Wu Y, Sun X. et al. Upregulation of MiR-212 Inhibits Migration and Tumorigenicity and Inactivates Wnt/β-Catenin Signaling in Human Hepatocellular Carcinoma. Technol Cancer Res Treat. 2018;17:1533034618765221
105. Zhang M, Li M, Li N, Zhang Z, Liu N, Han X. et al. miR-217 suppresses proliferation, migration, and invasion promoting apoptosis via targeting MTDH in hepatocellular carcinoma. Oncol Rep. 2017;37:1772-8
106. Dong Z, Qi R, Guo X, Zhao X, Li Y, Zeng Z. et al. MiR-223 modulates hepatocellular carcinoma cell proliferation through promoting apoptosis via the Rab1-mediated mTOR activation. Biochem Biophys Res Commun. 2017;483:630-7
107. Zhang C, Zhang J. Decreased expression of microRNA-223 promotes cell proliferation in hepatocellular carcinoma cells via the insulin-like growth factor-1 signaling pathway. Exp Ther Med. 2018;15:4325-31
108. Wang L, Bo X, Zheng Q, Xiao X, Wu L, Li B. miR-296 inhibits proliferation and induces apoptosis by targeting FGFR1 in human hepatocellular carcinoma. FEBS Lett. 2016;590:4252-62
109. Xie F, Yuan Y, Xie L, Ran P, Xiang X, Huang Q. et al. miRNA-320a inhibits tumor proliferation and invasion by targeting c-Myc in human hepatocellular carcinoma. Onco Targets Ther. 2017;10:885-94
110. Cui H, Song R, Wu J, Wang W, Chen X, Yin J. MicroRNA-337 regulates the PI3K/AKT and Wnt/β-catenin signaling pathways to inhibit hepatocellular carcinoma progression by targeting high-mobility group AT-hook 2. Am J Cancer Res. 2018;8:405-21
111. Liu P, Zhang H, Liang X, Ma H, Luan F, Wang B. et al. HBV preS2 promotes the expression of TAZ via miRNA-338-3p to enhance the tumorigenesis of hepatocellular carcinoma. Oncotarget. 2015;6:29048-50
112. Zhang T, Liu W, Zeng XC, Jiang N, Fu BS, Guo Y. et al. Down-regulation of microRNA-338-3p promoted angiogenesis in hepatocellular carcinoma. Biomed Pharmacother. 2016;84:583-91
113. Yuan J, Ji H, Xiao F, Lin Z, Zhao X, Wang Z. et al. MicroRNA-340 inhibits the proliferation and invasion of hepatocellular carcinoma cells by targeting JAK1. Biochem Biophys Res Commun. 2017;483:578-84
114. Yu M, Xue H, Wang Y, Shen Q, Jiang Q, Zhang X. et al. miR-345 inhibits tumor metastasis and EMT by targeting IRF1-mediated mTOR/STAT3/AKT pathway in hepatocellular carcinoma. Int J Oncol. 2017;50:975-83
115. Cui W, Li Y, Xu K, Chen G, Lu X, Duan Q. et al. miR-361-5p inhibits hepatocellular carcinoma cell proliferation and invasion by targeting VEGFA. Biochem Biophys Res Commun. 2016;479:901-6
116. Liu Y, Zhang W, Liu S, Liu K, Ji B, Wang Y. miR-365 targets ADAM10 and suppresses the cell growth and metastasis of hepatocellular carcinoma. Oncol Rep. 2017;37:1857-64
117. Xu J, Lin H, Li G, Sun Y, Chen J, Shi L. et al. The miR-367-3p increases sorafenib chemotherapy efficacy to suppress hepatocellular carcinoma metastasis through altering the androgen receptor signals. EBioMedicine. 2016;12:55-67
118. Pan XP, Wang HX, Tong DM, Li Y, Huang LH, Wang C. miRNA-370 acts as a tumor suppressor via the downregulation of PIM1 in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2017;21:1254-63
119. Xue H, Yu Z, Liu Y, Yuan W, Yang T, You J. et al. Delivery of miR-375 and doxorubicin hydrochloride by lipid-coated hollow mesoporous silica nanoparticles to overcome multiple drug resistance in hepatocellular carcinoma. Int J Nanomedicine. 2017;12:5271-87
120. Ge H, Zou D, Wang Y, Jiang H, Wang L. MicroRNA-377 Downregulates Bcl-xL and Increases Apoptosis in Hepatocellular Carcinoma Cells. Oncol Res. 2017;25:29-34
121. Xue H, Tian GY. MiR-429 regulates the metastasis and EMT of HCC cells through targeting RAB23. Arch Biochem Biophys. 2018;637:48-55
122. Liu X, Zhang A, Xiang J, Lv Y, Zhang X. miR-451 acts as a suppressor of angiogenesis in hepatocellular carcinoma by targeting the IL-6R-STAT3 pathway. Oncol Rep. 2016;36:1385-92
123. Zhao Y, Qi X, Chen J, Wei W, Yu C, Yan H. et al. The miR-491-3p/Sp3/ABCB1 axis attenuates multidrug resistance of hepatocellular carcinoma. Cancer Lett. 2017;408:102-11
124. Ye Y, Zhuang J, Wang G, He S, Zhang S, Wang G. et al. MicroRNA-495 suppresses cell proliferation and invasion of hepatocellular carcinoma by directly targeting insulin-like growth factor receptor-1. Exp Ther Med. 2018;15:1150-8
125. Yan JJ, Zhang YN, Liao JZ, Ke KP, Chang Y, Li PY. et al. MiR-497 suppresses angiogenesis and metastasis of hepatocellular carcinoma by inhibiting VEGFA and AEG-1. Oncotarget. 2015;6:29517-42
126. Wang D, Zhang N, Ye Y, Qian J, Zhu Y, Wang C. Role and mechanisms of microRNA-503 in drug resistance reversal in HepG2/ADM human hepatocellular carcinoma cells. Mol Med Rep. 2014;10:3268-74
127. Deng Q, Xie L, Li H. MiR-506 suppresses cell proliferation and tumor growth by targeting Rho-associated protein kinase 1 in hepatocellular carcinoma. Biochem Biophys Res Commun. 2015;467:921-7
128. Du X, Fan W, Chen Y. microRNA-520f inhibits hepatocellular carcinoma cell proliferation and invasion by targeting TM4SF1. Gene. 2018;657:30-8
129. Liu Y, Hong W, Zhou C, Jiang Z, Wang G, Wei G. et al. miR-539 inhibits FSCN1 expression and suppresses hepatocellular carcinoma migration and invasion. Oncol Rep. 2017;37:2593-602
130. Zhu C, Zhou R, Zhou Q, Chang Y, Jiang M. microRNA-539 suppresses tumor growth and tumorigenesis and overcomes arsenic trioxide resistance in hepatocellular carcinoma. Life Sci. 2016;166:34-40
131. Wu W, Dang S, Feng Q, Liang J, Wang Y, Fan N. MicroRNA-542-3p inhibits the growth of hepatocellular carcinoma cells by targeting FZD7/Wnt signaling pathway. Biochem Biophys Res Commun. 2017;482:100-5
132. Wang XP, Yao J, Guan J, Zhou ZQ, Zhang ZY, Yang J. MicroRNA-542-3p functions as a tumor suppressor via directly targeting survivin in hepatocellular carcinoma. Biomed Pharmacother. 2018;99:817-24
133. Jiang X, Wu J, Zhang Y, Wang S, Yu X, Li R. et al. MiR-613 functions as tumor suppressor in hepatocellular carcinoma by targeting YWHAZ. Gene. 2018;659:168-74
134. Zhang CZ, Cao Y, Fu J, Yun JP, Zhang MF. miR-634 exhibits anti-tumor activities toward hepatocellular carcinoma via Rab1A and DHX33. Mol Oncol. 2016;10:1532-41
135. Cheng J, Chen Y, Zhao P, Liu X, Dong J, Li J. et al. Downregulation of miRNA-638 promotes angiogenesis and growth of hepatocellular carcinoma by targeting VEGF. Oncotarget. 2016;7:30702-11
136. Zhang Y, Zhang D, Jiang J, Dong L. Loss of miR-638 promotes invasion and epithelial-mesenchymal transition by targeting SOX2 in hepatocellular carcinoma. Oncol Rep. 2017;37:323-32
137. Huang W, Li J, Guo X, Zhao Y, Yuan X. miR-663a inhibits hepatocellular carcinoma cell proliferation and invasion by targeting HMGA2. Biomed Pharmacother. 2016;81:431-8
138. Li Q, Li S, Wu Y, Gao F. miRNA-708 functions as a tumour suppressor in hepatocellular carcinoma by targeting SMAD3. Oncol Lett. 2017;14:2552-8
139. Zhang Y, Wei Y, Li X, Liang X, Wang L, Song J. et al. microRNA-874 suppresses tumor proliferation and metastasis in hepatocellular carcinoma by targeting the DOR/EGFR/ERK pathway. Cell Death Dis. 2018;9:130
140. Leong KW, Cheng CW, Wong CM, Ng IO, Kwong YL, Tse E. miR-874-3p is down-regulated in hepatocellular carcinoma and negatively regulates PIN1 expression. Oncotarget. 2017;8:11343-55
141. Xu Q, Zhu Q, Zhou Z, Wang Y, Liu X, Yin G. et al. MicroRNA-876-5p inhibits epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma by targeting BCL6 corepressor like 1. Biomed Pharmacother. 2018;103:645-52
142. Ding D, Zhang Y, Yang R, Wang X, Ji X, Ji G. et al. miR-940 suppresses tumor cell invasion and migration via regulation of CXCR2 in hepatocellular carcinoma. Biomed Res Int. 2016;2016:7618342
143. Zhao G, Dong L, Shi H, Li X, Guo X, Wang J. MicroRNA-1207-5p inhibits hepatocellular carcinoma cell growth and invasion through the fatty acid synthase-mediated Akt/mTOR signalling pathway. Oncol Rep. 2016;36:1709-16
144. Lin MF, Yang YF, Peng ZP, Zhang MF, Liang JY, Chen W. et al. FOXK2, regulted by miR-1271-5p, promotes cell growth and indicates unfavorable prognosis in hepatocellular carcinoma. Int J Biochem Cell Biol. 2017;88:155-61
145. Zhu H, Wang G, Zhou X, Song X, Gao H, Ma C. et al. miR-1299 suppresses cell proliferation of hepatocellular carcinoma (HCC) by targeting CDK6. Biomed Pharmacother. 2016;83:792-7
146. Yang C, Xu Y, Cheng F, Hu Y, Yang S, Rao J. et al. miR-1301 inhibits hepatocellular carcinoma cell migration, invasion, and angiogenesis by decreasing Wnt/beta-catenin signaling through targeting BCL9. Cell Death Dis. 2017;8:e2999
147. Ruijtenberg S, van den Heuvel S. Coordinating cell proliferation and differentiation: Antagonism between cell cycle regulators and cell type-specific gene expression. Cell Cycle. 2016;15:196-212
148. Urrego D, Tomczak AP, Zahed F, Stuhmer W, Pardo LA. Potassium channels in cell cycle and cell proliferation. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130094
149. Mohamed MS, Bishr MK, Almutairi FM, Ali AG. Inhibitors of apoptosis: clinical implications in cancer. Apoptosis. 2017;22:1487-509
150. Davis CD, Emenaker NJ, Milner JA. Cellular proliferation, apoptosis and angiogenesis: molecular targets for nutritional preemption of cancer. Semin Oncol. 2010;37:243-57
151. Matsuura K, Canfield K, Feng W, Kurokawa M. Metabolic regulation of apoptosis in cancer. Int Rev Cell Mol Biol. 2016;327:43-87
152. Jiang C, Li X, Zhao H, Liu H. Long non-coding RNAs: potential new biomarkers for predicting tumor invasion and metastasis. Mol Cancer. 2016;15:62
153. Yilmaz M, Christofori G, Lehembre F. Distinct mechanisms of tumor invasion and metastasis. Trends Mol Med. 2007;13:535-41
154. Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275-92
155. Zaravinos A. The regulatory role of micrornas in emt and cancer. J Oncol. 2015;2015:865816
156. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871-90
157. Yao H, Liu N, Lin MC, Zheng J. Positive feedback loop between cancer stem cells and angiogenesis in hepatocellular carcinoma. Cancer Lett. 2016;379:213-9
158. Terry K, Copur MS. Molecular targeted therapy of hepatocellular carcinoma. J Cancer Ther. 2013;4:426-39
159. Liu K, Min XL, Peng J, Yang K, Yang L, Zhang XM. The changes of HIF-1alpha and VEGF expression after TACE in patients with hepatocellular carcinoma. J Clin Med Res. 2016;8:297-302
160. Chen CK, Yu WH, Cheng TY, Chen MW, Su CY, Yang YC. et al. Inhibition of VEGF165/VEGFR2-dependent signaling by LECT2 suppresses hepatocellular carcinoma angiogenesis. Sci Rep. 2016;6:31398
161. Zhu AX, Duda DG, Sahani DV, Jain RK. HCC and angiogenesis: possible targets and future directions. Nat Rev Clin Oncol. 2011;8:292-301
162. Rahbari NN, Mehrabi A, Mollberg NM, Muller SA, Koch M, Buchler MW. et al. Hepatocellular carcinoma: current management and perspectives for the future. Ann Surg. 2011;253:453-69
163. Galun D, Srdic-Rajic T, Bogdanovic A, Loncar Z, Zuvela M. Targeted therapy and personalized medicine in hepatocellular carcinoma: drug resistance, mechanisms, and treatment strategies. J Hepatocell Carcinoma. 2017;4:93-103
164. Yuan P, Cao W, Zang Q, Li G, Guo X, Fan J. The HIF-2alpha-MALAT1-miR-216b axis regulates multi-drug resistance of hepatocellular carcinoma cells via modulating autophagy. Biochem Biophys Res Commun. 2016;478:1067-73
165. Chen KF, Chen HL, Tai WT, Feng WC, Hsu CH, Chen PJ. et al. Activation of phosphatidylinositol 3-kinase/Akt signaling pathway mediates acquired resistance to sorafenib in hepatocellular carcinoma cells. J Pharmacol Exp Ther. 2011;337:155-61
166. Aredia F, Scovassi AI. A new function for miRNAs as regulators of autophagy. Future Med Chem. 2017;9:25-36
167. Czaja MJ. Functions of autophagy in hepatic and pancreatic physiology and disease. Gastroenterology. 2011;140:1895-908
168. Chen L, Zhou Y, Sun Q, Zhou J, Pan H, Sui X. Regulation of autophagy by mirnas and their emerging roles in tumorigenesis and cancer treatment. Int Rev Cell Mol Biol. 2017;334:1-26
169. Lee YJ, Jang BK. The role of autophagy in hepatocellular carcinoma. Int J Mol Sci. 2015;16:26629-43
170. Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208-15
171. van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213-28
172. Tkach M, Thery C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell. 2016;164:1226-32
173. Li W, Li C, Zhou T, Liu X, Liu X, Li X. et al. Role of exosomal proteins in cancer diagnosis. Mol Cancer. 2017;16:145
174. Kosaka N. Decoding the Secret of Cancer by Means of Extracellular Vesicles. Journal of Clinical Medicine. 2016;5:22
175. Zhang L, Valencia CA, Dong B, Chen M, Guan PJ, Pan L. Transfer of microRNAs by extracellular membrane microvesicles: a nascent crosstalk model in tumor pathogenesis, especially tumor cell-microenvironment interactions. J Hematol Oncol. 2015;8:14
176. Zhang X, Yuan X, Shi H, Wu L, Qian H, Xu W. Exosomes in cancer: small particle, big player. J Hematol Oncol. 2015;8:83
177. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654-9
178. Zhang C, Ji Q, Yang Y, Li Q, Wang Z. Exosome: Function and Role in Cancer Metastasis and Drug Resistance. Technol Cancer Res Treat. 2018;17:1533033818763450
179. Pan JH, Zhou H, Zhao XX, Ding H, Li W, Qin L. et al. Role of exosomes and exosomal microRNAs in hepatocellular carcinoma: Potential in diagnosis and antitumour treatments (Review). Int J Mol Med. 2018;41:1809-16
180. Tang J, Li Y, Liu K, Zhu Q, Yang WH, Xiong LK. et al. Exosomal miR-9-3p suppresses HBGF-5 expression and is a functional biomarker in hepatocellular carcinoma. Minerva Medica. 2018;109:15-23
181. Wang H, Hou L, Li A, Duan Y, Gao H, Song X. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. Biomed Res Int. 2014;2014:864894
182. Liang G, Kan S, Zhu Y, Feng S, Feng W, Gao S. Engineered exosome-mediated delivery of functionally active miR-26a and its enhanced suppression effect in HepG2 cells. Int J Nanomedicine. 2018;13:585-99
183. Fu X, Liu M, Qu S, Ma J, Zhang Y, Shi T. et al. Exosomal microRNA-32-5p induces multidrug resistance in hepatocellular carcinoma via the PI3K/Akt pathway. J Exp Clin Cancer Res. 2018;37:52
184. Fang JH, Zhang ZJ, Shang LR, Luo YW, Lin Y, Yuan Y. et al. Hepatoma cell-secreted exosomal microRNA-103 increases vascular permeability and promotes metastasis by targeting junction proteins. Hepatology. 2018 [Epub ahead of print]
185. Lou G, Song X, Yang F, Wu S, Wang J, Chen Z. et al. Exosomes derived from miR-122-modified adipose tissue-derived MSCs increase chemosensitivity of hepatocellular carcinoma. J Hematol Oncol. 2015;8:122
186. Zhang Z, Li X, Sun W, Yue S, Yang J, Li J. et al. Loss of exosomal miR-320a from cancer-associated fibroblasts contributes to HCC proliferation and metastasis. Cancer Lett. 2017;397:33-42
187. Wang F, Li L, Piontek K, Sakaguchi M, Selaru FM. Exosome miR-335 as a novel therapeutic strategy in hepatocellular carcinoma. Hepatology. 2018;67:940-54
188. Shi M, Jiang Y, Yang L, Yan S, Wang YG, Lu XJ. Decreased levels of serum exosomal miR-638 predict poor prognosis in hepatocellular carcinoma. J Cell Biochem. 2018;119:4711-6
189. Sugimachi K, Matsumura T, Hirata H, Uchi R, Ueda M, Ueo H. et al. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br J Cancer. 2015;112:532-8
190. Fang T, Lv H, Lv G, Li T, Wang C, Han Q. et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun. 2018;9:191
191. Wang Y, Zhang C, Zhang P, Guo G, Jiang T, Zhao X. et al. Serum exosomal microRNAs combined with alpha-fetoprotein as diagnostic markers of hepatocellular carcinoma. Cancer Med. 2018;7:1670-9
192. Sohn W, Kim J, Kang SH, Yang SR, Cho JY, Cho HC. et al. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp Mol Med. 2015;47:e184
193. Liu WH, Ren LN, Wang X, Wang T, Zhang N, Gao Y. et al. Combination of exosomes and circulating microRNAs may serve as a promising tumor marker complementary to alpha-fetoprotein for early-stage hepatocellular carcinoma diagnosis in rats. J Cancer Res Clin Oncol. 2015;141:1767-78
194. Fornari F, Ferracin M, Trere D, Milazzo M, Marinelli S, Galassi M. et al. Circulating microRNAs, miR-939, miR-595, miR-519d and miR-494, Identify Cirrhotic Patients with HCC. PLoS One. 2015;10:e0141448
195. Yao DF, Dong ZZ, Yao M. Specific molecular markers in hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2007;6:241-7
196. He S, Zhang DC, Wei C. MicroRNAs as biomarkers for hepatocellular carcinoma diagnosis and prognosis. Clin Res Hepatol Gastroenterol. 2015;39:426-34
197. Shao H, Im H, Castro CM, Breakefield X, Weissleder R, Lee H. New Technologies for Analysis of Extracellular Vesicles. Chem Rev. 2018;118:1917-50
Author contact
![]() Corresponding authors: Prof. Yongchun Yu. No.274 Middle Zhijiang Road, Jingan District, Shanghai, P.R. China, 200071, Fax and Tel: 86-21-66313245, E-mail: yuyongchun1255com and Dr. Lifang Ma. No.274 Middle Zhijiang Road, Jingan District, Shanghai, P.R. China, 200071, Fax and Tel: 86-21-56634252, E-mail: malifang0606118com.
Corresponding authors: Prof. Yongchun Yu. No.274 Middle Zhijiang Road, Jingan District, Shanghai, P.R. China, 200071, Fax and Tel: 86-21-66313245, E-mail: yuyongchun1255com and Dr. Lifang Ma. No.274 Middle Zhijiang Road, Jingan District, Shanghai, P.R. China, 200071, Fax and Tel: 86-21-56634252, E-mail: malifang0606118com.

 Global reach, higher impact
Global reach, higher impact