3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(18):3269-3277. doi:10.7150/jca.25902 This issue Cite
Research Paper
The effect of Hsa_circ_0001451 in clear cell renal cell carcinoma cells and its relationship with clinicopathological features
1. Department of Urology, The First Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang Province, China
2. Department of Cardiology, KaiFeng Central Hospital, KaiFeng, Henan Province, China
3. Department of Neurosurgery, The First Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang Province, China
Received 2018-3-6; Accepted 2018-8-8; Published 2018-9-7
Abstract
Purpose: Circular RNAs (circRNAs), are a large class of RNAs that from a covalently closed continuous loop and have recently showed huge capabilities as gene regulators in mammals. Although Hsa_circ_0001451 has been investigated in colorectal cancer, it remains unclear about the relationship between Hsa_circ_0001451 and clear cell renal cell carcinoma (ccRCC). Our research aims to reveal the function of Hsa_circ_0001451 in the proliferation and development in ccRCC cells.
Methods: The expression of Hsa_circ_0001451 in 52 pairs of ccRCC tissues and paraneoplastic tissues was detected by quantitative real-time polymerase chain reaction (qRT-PCR). The correlation between Hsa_circ_0001451 and the clinicopathological features was evaluated using the chi-sequare test. Receiver operating characteristic (ROC) curve was built by SPSS to evaluate the diagnostic values. The effects of Hsa_circ_0001451 on ccRCC cells were determined via a MTT assay, clone formation assay, flow cytometry and Western blot analysis.
Results: The expression of Hsa_circ_0001451 was significantly correlated with differentiation (P<0.05). The area under ROC curve of Hsa_circ_0001451 was 0.704 (P<0.05). Knockdown of Hsa_circ_0001451 significantly promoted tumor growth in vitro. Bioinformatics results also displayed that Hsa_circ_0001451 might be involved in the regulation of tumor progression.
Conclusion: Taken together, our finding showed that Hsa_circ_0001451 might become a novel potential biomarker in the diagnosis of ccRCC and a potential novel target for the treatment of ccRCC.
Keywords: Circular RNA, Hsa_circ_0001451, clear cell renal cell carcinoma, biomarker, differentiation
Introduction
It is well known that renal cell carcinoma (RCC) is viewed as one of the most common urological tumors, which gradually has risen to the ninth leading cause of cancers in the US [1,2]. Among the three major subtypes of RCC, approximately 95% are clear cell RCC (ccRCC), of which the total survival rate of five years is about 74% [3,4]. Some recent studies have revealed that age of patients, tumor grade, and TNM stage are both closely related to the prognosis of RCC [5,6]. Recently, mutations in PBRM1, BAP1 and SETD2 are identified to be the molecular biomarkers for the prognosis of RCC [7, 8]. Molecular biomarkers may not only predict prognoses but also provide useful tumorigenic characteristics for the development of novel anti-RCC therapies.
Circular RNAs (circRNAs) represent a large class of non-coding RNAs that exist widespread on the cytoplasm of eukaryotic cells [9,10], which often are characterized as a stable structure and high tissue specific expression [11]. Compared with linear RNAs, circRNAs exhibit the remarkable characteristic of undergoing non-canonical splicing without 5'terminal cap and 3' terminal poly (A) tail and forming a ring structure with covalent bonds [12,13]. Recently, it was reported that circRNAs function as miRNA 'sponges' that naturally sequester and competitively suppress miRNA activity [14]. Furthermore, it was demonstrated that circRNAs were involved in the development of several types of disease, such as atherosclerosis and nervous system disorders [15,16]. However, the role of circRNAs in pathogenesis of renal cell carcinoma is still unknown.
In this study, we found that Hsa_circ_0001451 was downregulated both in renal cell carcinoma tissues and cells, and functioned as an anti-oncogene in renal cell carcinoma. SiRNA sequences targeting Hsa_circ_0001451 suppressed the expression of Hsa_circ_0001451 and promoted the progression of renal cell carcinoma.
Materials and Methods
Patients and clinical information
A total of 52 pairs of ccRCC tissue and adjacent normal tissue was collected from ccRCC patients who underwent radical nephrectomy at the First Affiliated Hospital of Harbin Medical University. All the samples were immediately snap frozen in liquid nitrogen for long-term preservation until RNA extraction.
Cell culture and siRNA transfection
OS-RC-2 and 786-O cells were grown in RPMI 1640 medium (KeyGen, Nanjing, China) and 10% FBS (Life Technologies, Australia). All cells were grown in at 37 °C in a humidified 5% CO2 atmosphere.
The OS-RC-2 and 786-O cells were seeded at six-well plates and then transfected at 24 h with specific siRNA (100 NM) or control siRNA (100 NM) using X-tremeGene according to the manufacturer's protocol (Roche). We used the following siRNA sequences: siRNA for Hsa_circ_0001451, sense: 5' CAACAAAAGAUUACUUCCUTT-3'; antisense: 5'AGGAAGUAAUCUUUUGUUGTT-3'.
Cell proliferation assay
Cell proliferation was assayed using the MTT assay. The transfected cells were plated in 96-well plates (5000cells/well). Cell proliferation was detected every 24 h according to the manufacturer's protocol. Briefly, 20 µl of MTT solution was added to each well and incubated for 4 h at 37 °C. The solution was then discard and add 150 µl of DMSO, after shaking 10 min at room temperature, the solution measured spectro photometrically at 450 nm.
Total RNA extraction and Quantitative real-time PCR
Total RNA was extracted from the ccRCC tissues and cell lines, using the TRIzol reagent (Ambion, life technologies, USA) according to the manufacturer's instruction. Total RNA was reverse transcribed into cDNA using the First Strand cDNA Synthesis Kit for detection of Hsa_circ_0001451 mRNA. The relative expression of Hsa_circ_0001451 mRNA was detected using FastStart Universal SYBR Green Master(ROX) and normalized to GAPDH. The primer sequences were as follows: 5'-CCAGTAGTATTGTGGACCTGCCC-3' (sense) and 5'-CCCTCTGACCCAGTAACTCCACT-3' (antisense) for Hsa_circ_0001451; 5'-CACCCACTCCTCCACCTTTGA-3' (sense) and 5'-ACCACCCTGTTGCTGTAGCCA-3' (antisense) for GAPDH. The result was analyzed by using -ΔCt method. All the results were expressed as the mean ± SD of three independent experiments.
Colony formation assay
OS-RC-2 and 786-O cells (0.1 × 103 cells per well) were seeded in a six-well plate and cultured for 10 days after treatment. Colonies were then fixed with 4% paraformaldehyde for 10 min and stained for 5 min with 0.5% crystal violet. Then the number of colonies was counted using ImageJ and images were taken under Olympus microscope (Tokyo, Japan).
Flow cytometry analysis of apoptosis
After transfection, the apoptosis level of OS-RC-2 and 786-O cell was analyzed using flow cytometry. The cells were collected and washed two times with PBS. Subsequently, the cells were stained by Annexin V-FITC and PI dye for 15 min in dark according to the manufacturer's instructions. The stained cells were analyzed by flow cytometry (BD Biosciences) and were counted using the CellQuest software (BD Biosciences). This experiment was repeated three times.
Hoechst staining
After transfection, cells were induced apoptosis using H2O2(100 µM for 2h),then cells were fixed 10min with the fixed solution, then washed it with PBS for 2 times, 3min each time and drained the liquid, Finally added 0.5ml Hoechst staining solution (Wanlei biological science and technology, Shenyang, China) to dye 5min. Images were taken under Olympus microscope (Tokyo, Japan).
Western blot analysis
Total proteins were prepared from OS-RC-2 and 786-O cells using RIPA buffer with proteinase inhibitor. The lysates were incubated on ice for 30 min, then centrifuged at 12,000 rpm for 15 min at 4 °C and protein concentration was measured by BCA kit (Beyotime Biotechnology, Beijing, China). Equal quantities of protein were electrophoresed through a 12.5% sodium dodecyl sulfate/poly-acrylamide gel and transferred to nitrocellulose membranes (Millipore, Billerica, MA). The membranes were blocked and then incubated with Bax, Bcl-2, and β-actin overnight at 4 °C. Subsequently, the membranes were incubated with the anti-mouse or rabbit secondary antibody (Santa Cruz Biotechnology) at room temperature for 1h. The protein bands were visualized using a chemiluminescence reagent (ECL) kit (Beyotime Biotechnology).
Functional enrichment analyses
To evaluate the functional implication of Hsa_circ_0001451, Circular RNA Interactome was used to predict the target miRNA and starBase was used to predict the miRNA targeted mRNA. The ceRNA network was visualized using Cytoscape software (version 3.4.0) [17]. Functional enrichment analyses for those predict genes were performed using the DAVID Bioinformatics Tool (version 6.7) [18]. GO and KEGG category enrichments were based on the threshold of P-value < 0.05 and enrichment score > 1.0. Significant enrichment results were visualized using R language.
Statistical analysis
Statistical data were analyzed using Statistical Program for Social Sciences (SPSS) 19.0 software (SPSS, Chicago, IL, USA). GraphPad Prism7.0 (GraphPad Software, La Jolla, CA) was used to plot all graphs. The differences between tumor tissues and normal colon mucosa were analyzed by paired t-test. Receiver operating characteristic (ROC) curve was constructed to evaluate the diagnostic values. P<0.05 was considered statistically significant.
Results
Hsa_circ_0001451 is significantly downregulation in ccRCC tissues
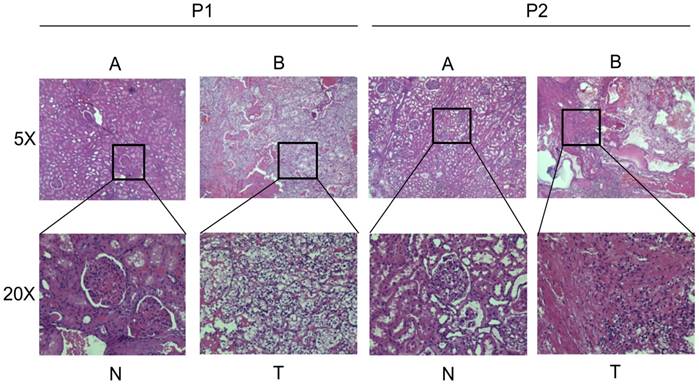
In order to exclude the possible influence of cancer types, we just selected patients diagnosed with clear cell renal cell carcinoma by HE stain. Figure 1A shows the normal kidney tissue (HE, ×50; HE, ×200), and Figure 1B shows the clear cell renal cell carcinoma tissue (HE, ×50; HE, ×200). To make clear about the potential significance of Hsa_circ_0001451 in the progression of ccRCC, RT-qPCR was applied to measure the level of Hsa_circ_0001451 in ccRCC tissues and cells. As shown in Fig 2A, the expression level of Hsa_circ_0001451 was significantly downregulated in ccRCC tissues compared with the matched normal tissues (p < 0.001). These data implied that Hsa_circ_0001451 might be associated with the progression of ccRCC.
General Pathology of the patients. (A) The normal kidney tissue (HE, ×50; HE, ×200). (B) The clear cell renal cell carcinoma tissue (HE, ×50; HE, ×200)
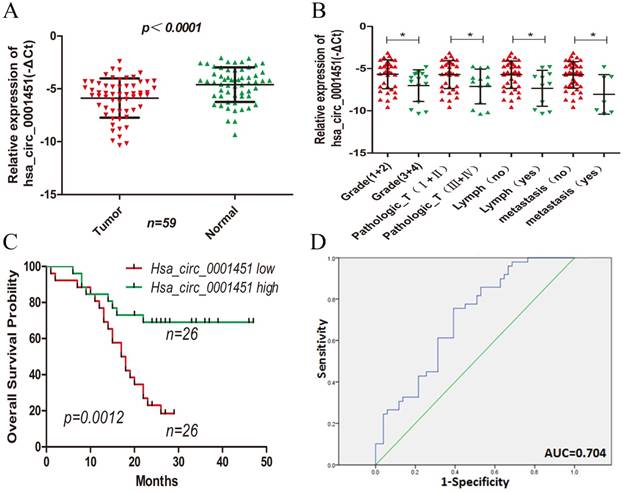
Hsa_circ_0001451 expression is downregulated in ccRCC tissues and its clinical significance. (A) Relative expression of Hsa_circ_0001451 in human ccRCC tissues (n=28) compared with corresponding adjacent normal tissues (n=28). Hsa_circ_0001451 expression was examined by qPCR and normalized to GAPDH expression (shown as - ΔCT). (B) The patients were classified into two groups according to Hsa_circ_0001451 expression. The results are presented as relative expression levels in tumor tissues. Hsa_circ_0001451 expression was significantly lower in patients with a higher grade, a higher pathological stage, lymph node metastasis and distant metastasis (shown as -ΔCT). Bars: s.d, *P<0.05. (C) The relationship between low levels of Hsa_circ_0001451 and overall survival was analyzed by Kaplan-Meier survival analysis.(D) The ROC curve was constructed by SPSS. Area under curve was 0.704. The sensitivity and specificity was 0.755 and 0.608, respectively. P<0.05.
The relationship of hsa_circ_0001451 expression levels (-ΔCt) in cancer tissue with clinical features in KIRC patients
| Variable | KIRC | Group | p-Value | |
|---|---|---|---|---|
| Low circ | High circ | |||
| Sample (n) | 52 | 26 | 26 | - |
| Median circ | - -6.0089 (-2.39--10.3596) | - | - | - |
| Median age (year) | 56.8(35-82) | - | -- | 0.402 |
| ≤ 57 | 29(55.8%) | 16 | 13 | |
| >57 | 23(44.2%) | 10 | 13 | |
| Gender | 0.777 | |||
| Male | 31(59.6%) | 15 | 16 | |
| Female | 21(40.4%) | 11 | 10 | |
| Clinical stage | 0.006* | |||
| Stage I+ II | 37(71.2%) | 14 | 23 | |
| Stage III+ IV | 15(28.8%) | 12 | 3 | |
| Tumor stage | 0.025* | |||
| T1+ T2 | 39(75.0%) | 16 | 23 | |
| T3+ T4 | 13(25.5%) | 10 | 3 | |
| Lymph node metastasis | 0.017* | |||
| N0 | 41(78.8%) | 17 | 24 | |
| N1 | 11(21.2%) | 9 | 2 | |
| NX | - | - | - | |
| Distant metastasis | 0.042* | |||
| M0 | 45(86.5%) | 20 | 25 | |
| M1 | 7(13.5%) | 6 | 1 | |
| MX | - | - | - | |
| Staus | 0.000* | |||
| Dead | 29(55.8%) | 21 | 8 | |
| Alive | 23(44.2) | 5 | 18 | |
Low/high by the sample mean. Pearson χ2-test. p < 0.05 was considered statistically significant
Hsa_circ_0001451 is correlated with the clinicopathological features and overall survival of ccRCC patients
Next, we analyzed the correlation between the expression of Hsa_circ_0001451 with the clinicopathological parameters of 52 patients definitely diagnosed with ccRCC. The mean expression level of Hsa_circ_0001451 in all ccRCC tissues was set as a cutoff value, and all samples were fallen into two groups (highly expressed group n = 26 vs. under expressed group n = 26). We found that the expression of Hsa_circ_0001451 was related to clinical stage, tumor stage, lymph node and metastasis, rather than gender and age of patients (Table 1; Figure 2B). Kaplan-Meier survival analysis disclosed that low level of Hsa_circ_0001451 was negatively correlated with overall survival of patients with ccRCC (Fig. 2C, p = 0.0012). Then we included the variable including age, gender, clinical stage, tumor stage, lymph node, metastasis, and Hsa_circ_0001451 mRNA level into a multivariate Cox regression model and found that the mRNA level of Hsa_circ_0001451 was an independent predictor for overall status of ccRCC patients (HR=0.375, 95%CI 0.142-0.987, p=0.047, Table 2). These results suggested a link between down-regulation of Hsa_circ_0001451 and aggressive charateristics of ccRCC might act as an anti-oncogene in ccRCC.
Hsa_circ_0001451 could be used as indictor of ccRCC
To assessment whether Hsa_circ_0091451 could be used as an indicator for diagnosis of ccRCC, a ROC curve was built. A total of 52 patients' normal kidney tissues were used as a control to produce this ROC curve (Figure 2D). The sensitivity and specificity were 0.755 and 0.392, respectively. The cutoff value was -4.701. The area under the curve was 0.704 (95% CI=0.603-0.805, P<0.000). The Youden index was 0.363. Therefore, Hsa_circ_0001451 could be used as indictor of ccRCC.
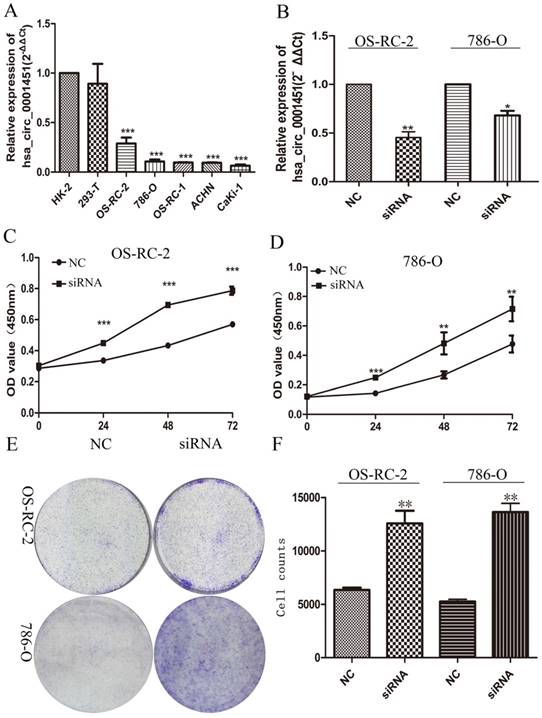
Knockdown of Hsa_circ_0001451 promotes proliferation of ccRCC cell line in vitro
To further investigate the function of Hsa_circ_0001451, we firstly detected its expression level in the renal cell carcinoma cell lines (OS-RC-2,786-O, OS-RC-1, ACHN, Caki1) and the normal cell line (HK-2, 293-T), respectively. All cancer cell lines showed pronounced low expression compared with normal cell line (Figure 3A). Then we designed siRNA to inhibit Hsa_circ_0001451 and came to test its biological function in vitro. The cell lines OS-RC-2 and 786-O were transfected with si-Hsa_circ_0001451 or negative control (NC). 48 h after treatment, the expression levels of Hsa_circ_0001451 were effectively decreased (Fig. 3B). The MTT assay results showed that knockdown of Hsa_circ_0001451 significantly promoted the proliferation of OS-RC-2 and 786-O cell lines (Fig. 3C and D). Furthermore, the clone formation experiment also showed that si-Hsa_circ_0001451 promoted cell proliferation (Fig. 3E and F). The findings above indicated that knockdown of hsa_circ_000145 could enhance the proliferation of ccRCC cells.
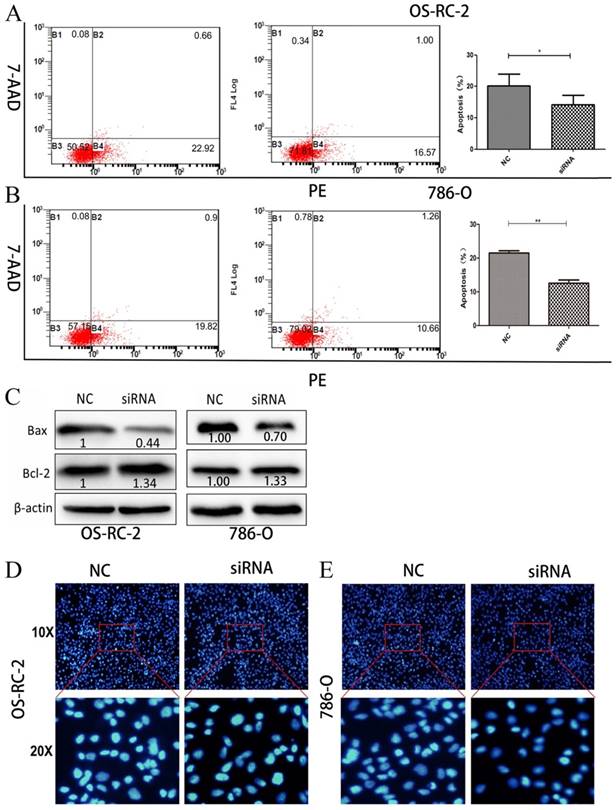
Knockdown of Hsa_circ_0001451 can inhibition of cell apoptosis
We then performed flow cytometric analyses to further evaluate whether Hsa_circ_0001451 affected proliferation of OS-RC-2 and 786-O cells by altering apoptosis. OS-RC-2 and 786-O cells transfected with si-Hsa_circ_0001451 obviously inhibited cell apoptosis (Fig. 4A and B). In order to verify the cause of cell apoptosis change, the Bax and Bcl-2 protein levels were assessed in OS-RC-2 and 786-O by western blot after si-Hsa_circ_0001451 or si-NC transfection (Fig. 4C). Hoechst staining assays further confirmed the results (Fig. 4D and E). Our results suggested that inhibition of Hsa_circ_0001451 promoted the proliferation of OS-RC-2 and 786-O cells and affected cell apoptosis.
Discussion
Recently, Circular RNAs (circRNAs) are discovered as new endogenous non-coding RNAs [19,20,21]. In structure, compared with the traditional linear RNA (linear RNA, 5' and 3' ends) [22,23], circRNA molecule is a closed ring structure, so that it is not affected by exonuclease, making it very stable and difficult to degrade the expression [24,25]. In function, recent studies have shown that circRNA molecule contain microRNA (miRNA) binding sites to miRNA sponge in the cell function [26,27], thus lead to raise the inhibitory effect of miRNA on its target gene and increase expression of target genes, this mechanism is called competitive endogenous RNA (ceRNA) mechanism [28,29]. By interacting with miRNA, circRNA plays an important regulatory role in disease, especially in cancer [30]. In research of ccRCC, many studies have focused on the epigenetic regulation of its pathogenesis and potential targets for therapy, including microRNAs and long noncoding RNAs (lncRNAs). However, the occurrence of circRNAs in cancer remains largely unknown. This study aims to expound circRNA expression in human kidney tissue. In the process of research, we identified that Hsa_circ_0001451 is aberrantly expressed in ccRCC compared with normal kidney tissue. Through loss of function, we found that Hsa_circ_0001451 is important in the process of the development of clear cell renal cell carcinoma.
Univariable and multivariable Cox regression analyses in KIRC.
| Characteristics | Subset | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | ||
| Age | ≤ 57/>57 | 1.579 (0.760-3.277) | 0.221 | 1.187(0.492-2.864) | 0.704 |
| Gender | Male/Female | 1.067 (0.509-2.236) | 0.864 | 1.529(0.680-3.437) | 0.304 |
| Pathologic_T | Ⅰ+Ⅱ/Ⅲ+Ⅳ | 4.759 (2.243-10.097) | 0.000* | 4.660(0.732-29.652) | 0.103 |
| Grade | Grade (1+2)/Grade (3+4) | 3.019 (1.440-6.329) | 0.003* | 0.658(0.153-2.841) | 0.575 |
| Lymph node metastasis | Yes/No | 4.094 (1.907-8.786) | 0.000* | 1.209(0.249-5.863) | 0.814 |
| Distant metastasis | Yes/No | 3.298 (1.397-7.787) | 0.006* | 0.692(0.195-2.459) | 0.569 |
| Expression | High/Low | 0.283 (0.124-0.642) | 0.003* | 0.375(0.142-0.987) | 0.047* |
In both univariable and multivariable Cox regression analyses, age, gender, Pathologic_T and Tumor grade, Lymph node metastasis, Distant metastasis, Expression were evaluated as continuous variables. P<0.05 was considered statistically significant in all analyses.
Hsa_circ_0001451 knockdown promote ccRCC cell proliferation in vitro. (A) Hsa_circ_0001451 expression levels of RCC cell lines (OS-RC-2,786-O, OS-RC-1, ACHN, Caki-1), compared with that in human Renal tubular epithelial cells (HK-2,293-T,). (B) qRT-PCR analysis of Hsa_circ_0001451 expression in OS-RC-2 and 786-O cell lines transfected with Hsa_circ_0001451 siRNAs or the negative control. (C, D) MTT assays were performed to detect the viability of si-Hsa_circ_0001451 transfected OS-RC-2 and 786-O cell lines. (E, F) Colony-forming growth assays were performed to determine the proliferation of OS-RC-2 and 786-O cell lines. The colonies were counted and captured.
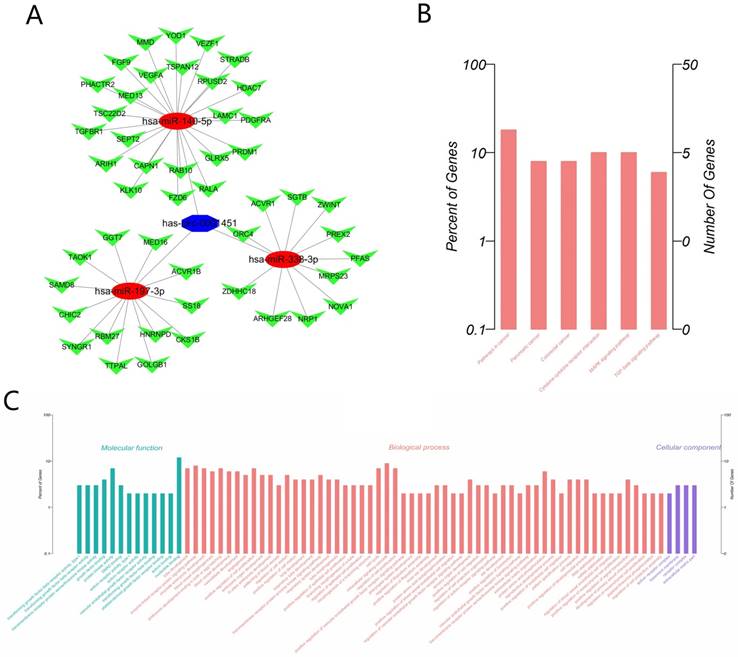
Considering that circRNAs often function as the form of molecular sponges, we predicted target miRNAs of Hsa_circ_0001451 through Circular RNA Interactome (https://circinteractome.nia.nih.gov/), and thereby got about 25 target miRNAs in all. Excluding those non-conservative miRNA and without TCGA data support, we finally determined three miRNAs (Hsa-miR-140-5p, Hsa-miR-197-3p, Hsa-miR-338-3p, Supplementary material, Table S1) for the next study. Then we predicted the target mRNA of the three miRNAs through starBase v2.0 (http://starbase.sysu.edu.cn/) under conditions of medium stringency and at list three prediction software support, and then compared with TCGA, 48 genes were collected (Figure 5A, Supplementary material Table S2). Gene Ontology (GO) enrichment analysis was applied to explore the function of targeted mRNAs identified in this study. Genes were organized into hierarchical categories to uncover gene regulatory networks on the basis of biological process, cellular component and molecular function. Specifically, two-side Fisher's exact test was used to determine the GO category and GO annotation list was greater than expected by chance (P value < 0.05 is recommended as the cut-off). Through GO analysis we found that these genes were associated with cell cycle (ontology: biological process), receptor complex (ontology: cellular component) and nucleotide binding (ontology: molecular function) (Figure 5C, Supplementary material, Table S3). Through the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, we identified that 6 pathways were significantly enriched, including Hsa05200: Pathways in cancer, Hsa05212: Pancreatic cancer, Hsa0210: Colorectal cancer, Hsa04060: Cytokine-cytokine receptor interaction, Hsa04010: MAPK signal pathway, Hsa04350: TGF-beta signaling pathway, but only the enrichments of Hsa05200: Pathways in cancer were significant after FDR correction (Figure 5B, Supplementary material, Table S4).
However, our study just validated the existence of Hsa_circ_0001451 in 52 patients. There is no doubt that in the future research, except needing more samples, it's necessary to explore more function of molecular mechanisms of Hsa_circ_0001451 in the progression of ccRCC.
In conclusion, Hsa_circ_0001451 could be detected as a circRNA in human renal cell carcinoma (RCC) tissues and adjacent tissues. It was found that the expression of Hsa_circ_0001451 was down-regulated in RCC tissues. Furthermore, its expression was associated with differentiation and invasion. These results suggested that Hsa_circ_0001451 might be a new target for treatment and a potential biomarker for RCC, providing a valuable basis for the future treatment of RCC.
Knockdown of Hsa_circ_0001451 inhibit apoptosis in OS-RC-2 and 786-O cells in vitro. (A, B) Flow cytometry was used to detect the apoptotic rates of cells. LR, early apoptotic cells; UR, terminal apoptotic cells. (C) Western blot analysis of Bax and Bcl-2 after si-NC, si- Hsa_circ_0001451 transfection in OS-RC-2 and 786-O cells. β-actin protein was used as an internal control.(D) Hoechst staining assay performed in OS-RC-2 and 786-O cells transfected with si- Hsa_circ_0001451 or si-NC.
The Biological function prediction of Hsa_circ_0001451 by Bioinformatics. (A) The ceRNA network of the predict miRNA and mRNA map. (B) KEGG pathways significantly associated with the predict protein-coding genes. (C) The functional enrichment map of the GO terms.
Supplementary Material
Supplementary tables.
Acknowledgements
This work was supported by National Natural Science Foundation of China [grant No.81171996; 81272289]. The First Affiliated Hospital of Harbin Medical University Science Foundation [grant No. 2015B011].
Author Contributions
Wang Gang mainly performed the experiments, analyzed the data and wrote the paper. Xue Wei and Jian Wengang helped with the experiments. Liu Panhong helped analyzed the data. Wang Zichun helped modify the paper. Wang Changlin performed MTT assay. Li Haoming performed western blot and prepared the samples. Yu Yipeng helped with the paper writing. Zhang Daming and Zhang Cheng carried out the experiment design and manuscript drafting. All authors had edit and approved the final manuscript.
Ethics approval and consent to participate
This study was conducted at Department of Urology, The First Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang, China. This research study was approved by the Institutional Review Board of Harbin Medical University, and the subjects provided informed consent.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. Ca A Cancer Journal for Clinicians. 2014;65:5
2. Turajlic S, Larkin J, Swanton C. Snapshot: Renal cell carcinoma. Cell. 2015;163:1556
3. Min-Han Tan MM, Huihua Li Ph D, Caroline Victoria Choong. et al. The karakiewicz nomogram is the most useful clinical predictor for survival outcomes in patients with localized renal cell carcinoma. Cancer. 2011;117:5314
4. Chow WH, Devesa SS, Warren JL. et al. Rising incidence of renal cell cancer in the united states. Jama. 1999;281:1628
5. Schmittgen TD, Livak KJ. Analyzing real-time pcr data by the comparative c(t) method. Nature Protocols. 2008;3:1101
6. Moch H, Gasser T, Amin MB. et al. Prognostic utility of the recently recommended histologic classification and revised tnm staging system of renal cell carcinoma: A swiss experience with 588 tumors. Cancer. 2000;89:604
7. Cocquerelle C, Mascrez B, Hétuin D. et al. Mis-splicing yields circular rna molecules. Faseb Journal Official Publication of the Federation of American Societies for Experimental Biology. 1993;7:155
8. Salzman J, Gawad C, Wang PL. et al. Circular rnas are the predominant transcript isoform from hundreds of human genes in diverse cell types. PloS one. 2012;7:e30733
9. Conn SJ, Pillman KA, Toubia J. et al. The rna binding protein quaking regulates formation of circrnas. Cell. 2015;160:1125
10. Kulcheski FR, Christoff AP, Margis R. Circular rnas are mirna sponges and can be used as a new class of biomarker. Journal of biotechnology. 2016;238:42-51
11. Hentze MW, Preiss T. Circular rnas: Splicing's enigma variations. Embo Journal. 2013;32:923-925
12. Vicens Q, Westhof E. Biogenesis of circular rnas. Cell. 2014;159:13-14
13. Hansen TB, Jensen TI, Clausen BH. et al. Natural rna circles function as efficient microrna sponges. Nature. 2013;495:384-388
14. Holdt LM, Anika S, Kristina S. et al. Circular non-coding rnaanrilmodulates ribosomal rna maturation and atherosclerosis in humans. Nature communications. 2016;7:12429
15. Liu Q, Zhang X, Hu X. et al. Circular rna related to the chondrocyte ecm regulates mmp13 expression by functioning as a mir-136 'sponge' in human cartilage degradation. Sci Rep. 2016;6:22572
16. You X, Vlatkovic I, Babic A. et al. Neural circular rnas are derived from synaptic genes and regulated by development and plasticity. Nature Neuroscience. 2015;18:603-610
17. Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009;37:1
18. Kulcheski FR, Christoff AP, Margis R. Circular rnas are mirna sponges and can be used as a new class of biomarker. Journal of biotechnology. 2016;238:42
19. Han D, Li J, Wang H. et al. Circular rna mto1 acts as the sponge of mir-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017
20. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498-2504
21. Wang K, Sun Y, Tao W. et al. Androgen receptor (ar) promotes clear cell renal cell carcinoma (ccrcc) migration and invasion via altering the circhiat1/mir-195-5p/29a-3p/29c-3p/cdc42 signals. Cancer letters. 2017;394:1-12
22. Floris G, Zhang L, Follesa P. et al. Regulatory role of circular rnas and neurological disorders. Molecular Neurobiology. 2017;54:5156-5165
23. Xia W, Qiu M, Chen R. et al. Circular rna has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Scientific Reports. 2016;6:35576
24. Conn SJ, Pillman KA, Toubia J. et al. The rna binding protein quaking regulates formation of circrnas. Cell. 2015;160:1125-1134
25. Zheng Q, Bao C, Guo W. et al. Circular rna profiling reveals an abundant circhipk3 that regulates cell growth by sponging multiple mirnas. Nature communications. 2016;7:11215
26. Liu Y, Cui H, Wang W. et al. Construction of circular mirna sponges targeting mir-21 or mir-221 and demonstration of their excellent anticancer effects on malignant melanoma cells. International Journal of Biochemistry & Cell Biology. 2013;45:2643-2650
27. Chen Y, Li C, Tan C. et al. Circular rnas: A new frontier in the study of human diseases. Journal of Medical Genetics. 2016;53:359
28. Barrett SP, Salzman J. Circular rnas: Analysis, expression and potential functions. Development. 2016;143:1838-1847
29. Li P, Chen H, Chen S. et al. Circular rna 0000096 affects cell growth and migration in gastric cancer. British Journal of Cancer. 2017;116:626
30. Qu S, Yang X, Li X. et al. Circular rna: A new star of noncoding rnas. Cancer letters. 2015;365:141-148
Author contact
![]() Corresponding authors: Cheng Zhang, Department of Urology, The First Affiliated Hospital of Harbin Medical University, Harbin 150001, Heilongjiang, China. Email: doctorcheng77com and Daming Zhang, Department of Neurosurgery, The First Affiliated Hospital of Harbin Medical University, Harbin 150001, Heilongjiang, China.Email: zhangdamingcom
Corresponding authors: Cheng Zhang, Department of Urology, The First Affiliated Hospital of Harbin Medical University, Harbin 150001, Heilongjiang, China. Email: doctorcheng77com and Daming Zhang, Department of Neurosurgery, The First Affiliated Hospital of Harbin Medical University, Harbin 150001, Heilongjiang, China.Email: zhangdamingcom

 Global reach, higher impact
Global reach, higher impact