3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(17):2994-3005. doi:10.7150/jca.25876 This issue Cite
Research Paper
Loss of UHRF2 Is Associated With Non-small Cell Lung Carcinoma Progression
1. Department of Thoracic Surgery, The Affiliated Zhongshan Hospital of Fudan University, Shanghai 200032, P. R. China
2. Department of Cardiothoracic Surgery, The Second Affiliated Hospital of Nanchang University, Jiangxi Province 330000, P. R. China.
3. Department of emergency surgery, Fujian Provincial Hospital, Fu Zhou, Fujian Province,350001, China
* These authors contributed equally to this work.
Received 2018-3-4; Accepted 2018-6-9; Published 2018-7-30
Abstract
Recent evidence indicated ubiquitin like with PHD and ring finger domains 2 (UHRF2) was involved in various human diseases, especially in cancer, however, its roles in cancer are still in dispute. Here, we found UHRF2 expression was decreased in lung cancer tissues compared with adjacent normal tissues by referring to the Oncomine Database, which was further identified by immunoblotting and quantitative real-time polymerase chain reaction assays. Secondly, we found knockdown of UHRF2 in A549 and 95-D cell lines enhanced the capability of proliferation, invasion and migration, while forced UHRF2 expression inhibited NSCLC cells proliferation,invasion and migration. Mechanistically, dot-blot and western blot assays indicated that the level of UHRF2 was positively correlated with 5-hmC level by affecting ten-eleven translocation 2 (TET2) expression. Clinically, UHRF2 downregulation is significantly correlated with a malignant phenotype, including larger tumor size and poor differentiation. Moreover, UHRF2 downregulated correlates with shorter overall survival(OS).
Conclusion: Our findings indicate that UHRF2 is a tumor suppressor in NSCLC by influence TET2 expression and serve as a potential therapeutic target in NSCLC.
Keywords: NSCLC, UHRF2, 5-hmC, demethylation
Introduction
Lung cancer is one of the major types of cancer and the leading cause of cancer deaths worldwide. In 2017, there were estimated 0.22 million new cases and estimated 0.15 million deaths in United States [1]. Clinically, lung cancer is classified as small-cell lung carcinoma (SCLC) and non-small cell lung carcinoma (NSCLC), and NSCLC accounts for nearly 85% of lung cancer cases. Despite the advancement in diagnostic approaches and introduction of new therapeutic procedures over past decades [2], the 5-year survival rate of NSCLC remains dismal due to early metastasis and high recurrence rate. Thus, the identification of new biomarkers that shape cancer progression will be assisted with probing for effective prognostic markers and more effective therapeutic targets to improve the outcome of NSCLC patients.
UHRF2 (ubiquitin like with PHD and ring finger domains 2) includes UBL, PHD, TTD, SET and RING-associated (SRA/YDG) and RING finger domains [3]. Due to its multiple domains, UHRF2 has a complex function, including as a cell cycle regulator, regulating genomic stability and epigenetics [4]. Recently, some studies reported UHRF2 was involved in various human diseases, especially in cancer, however, its exact roles in cancer are still in dispute. For example, it was reported that UHRF2 contributed to the progression of colon carcinogenesis [5], while others argued that UHRF2 was a tumor supressor in leukemia [6, 7]. Thus, the function of UHRF2 is still ill-defined, and further study is needed.
DNA methylation is an important epigenetic modification that plays a role in diverse biological processes, including maintenance of histone modification, genomic stability, and chromatin remodelling [8, 9]. Genomic DNA 5-methylcytosine hydroxylation (5-hmC) is considered to be a demethylation intermediates status which is distributed unevenly across different tissues and cells [10]. The levels of 5-hmC are generally low in cancer cells and cultured somatic cells, but are relatively high in pluripotent or multipotent cells, such as ESCs (embryonic stem cells) or neural stem cells [11]. 5-hmC also show specific disease-specific changes in the circulating cell-free DNA of different types of cancer. Moreover, loss of 5-hmC was reported to be a prognostic marker and an oncogenic event in several cancers [12]. In total, 5-hmC is closly associated with the development of cancer.
Recently, UHRF2 is identified as a reader of 5-hmC [13], suggesting its specific biological roles on demethylation process. Here, we tried to reveal the correlation between UHRF2 and 5-hmC and the role of UHRF2 in NSCLC.
Method and Materials
Patients and specimens
Archival specimens were obtained from 208 patients at Zhongshan Hospital (Shanghai, People's Republic of China) in 2005 with informed consent. All the patients who underwent curative resection for NSCLC were included in our study. All patients underwent standard lobectomy and mediastinal lymph node dissection. Paraffin blocks were selected only on the basis of the availability of suitable formalin-fixed, paraffin-embedded tissue and complete clinicopathologic and follow-up data for the patients. Briefly, tumor stage was adjudged on the basis of the tumor-node-metastasis (TNM) 7th edition of International Union Against Cancer Staging Manual. Pathological classification was based on World Health Organization criteria. Follow-up was completed in July 2010. The median follow-up was 43 months (range, 1-66 months). Overall survival (OS) was defined as the interval between surgery and death or between surgery and the last observation for surviving patients. The data were censored at the last follow-up for living patients. Sixteen fresh NSCLC and their adjacent normal lung samples were obtained from other NSCLC patients. Ethical approval was obtained from the Zhongshan Hospital Research Ethics Committee.
Cell Lines and cell cultures
The NSCLC lines 95-D, and A549 were purchased from the Institute of Biochemistry and Cell Biology at the Chinese Academy of Science [14]. The cells were maintained in Dulbecco modified Eagle medium or RPMI 1640 supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 0.1 mg/ml streptomycin.
Tissue Microarrays and Immunohistochemistry
All samples from NSCLC patients were reviewed histologically by hematoxylin and eosin staining, and representative areas with small round lymphocyte infiltrate were premarked in the paraffin blocks, away from necrotic and hemorrhagic materials. Duplicates of 1-mm-diameter cylinders from two different areas, tumor center and nearest noncancerous margin (designated as intratumor and peritumor, respectively; a total of four punches) were included in each case, along with different controls, to ensure reproducibility and homogenous staining of the slides (Shanghai Biochip Co.Ltd., Shanghai, People's Republic of China). Thus, four different tissue microarray blocks were constructed, each containing 312 cylinders. Sections 4µm thick were placed on slides coated with 3-aminopropyltriethoxysilane. UHRF2 polyclonal antibodies (abcam, ab28673) and 5hMC antibodies (Active Motif, 39791) were used to detect the expression of their respective proteins. The UHRF2 density was counted by Image-Pro Plus v6.2 software (Media Cybernetics, Inc., Bethesda, MD). For the reading of each antibody staining, a uniform setting for all the slides was applied. Integrated optical density (IOD) of all positive staining of UHRF2 in each photograph was measured and normalized to the mean value of all photograph. The intensity of UHRF2 was classified into high UHRF2 expression (UHRF2+), and low UHRF2 expression (UHRF2-). The same method above was applied to 5-hmC staining.
Plasmids and Retroviral/Lentiviral Constructs and Transduction
The plasmid MigR1-UHRF2 were purchased from General Biosystems (Anhui) Co Ltd. MigR1 constructs and the plasmids psRa-g (encoding envelope) and PMDold gag-pol (encoding gag-pol) were introduced into 293T cells by co-transfection using Lipofectamine 2000 (Life Technologies). 48 hours later, the supernatants containing virus were harvested for future assays.
shRNA sequences were synthesized by Shanghai HuaGene Biotechnology Co. Ltd., and the oligos were cloned into the shRNA lentiviral vector PLKO.1. 293T cells were were transfected using Lipofectamine 2000 (Life Technologies) with 2 µg of PLKO.1-shCK or PLKO.1-shRNA1/shRNA2, together with 0.5 µg of VSV-g and 1.5 µg Pax2 viral plasmids. 48 hours later, the supernatants containing virus were harvested for future knockdown assays. A549 and 95-D were incubated with viral supernatants containing 6µg/ml of Polybrene overnight. The viral supernatants were replaced with fresh medium on day 2. Puromycin was added to screen the cells 2 days post-transduction for stable cell lines. Stably transfected cells were validated by qRT-PCR and IB. The shRNA sequences were as follows: CAACAAGATGAAGAGCACCAA(shCK), CGTCTCTTCTTCCATTACAAT(UHRF2-shRNA1), AGTGTACCCTCTACGTCTAAT(UHRF2-shRNA2). The correct sequence of all plasmids was verified by sequencing (sequences of plasmids and oligonucleotides on request).
Genomic DNA isolation and Dot blot assay
A549 and 95-D genomic DNA was extracted with the Blood & Cell Culture DNA Mini Kit (QIAGEN), The DNA concentration was measured by NanoDrop (Thermo Scientific). Briefly, the procedures of dot blot as follows, DNA was spotted on a nitrocellulose membrane (Whatman), which was placed under an ultraviolet lamp for 20 min to crosslink the DNA. Subsequently, the membrane was blocked with 5% milk in TBS-Tween for 1 h and incubated with the primary anti-5hMC antibodies (Active Motif, 39791) at 4°C overnight. After incubation with a horseradish peroxidase-conjugated secondary antibody anti-rabbit IgG (GenScript) for 1 h at room temperature, the membrane was washed with PBS-Tween 20 for three times and then detected and scanned by a Tanon6100 scanner (Tanon,China). The 5hmC intensity was quantified by Image-J software (USA).
Immunoblotting and Quantitative Real-Time Polymerase Chain Reaction analysis
Cells were seeded, transfected in six-well plates (Corning). After 48 or 72 h, cells were harvested, washed once with PBS and the pellets lysed in buffer consisting of 20 nM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1%Nonidet P-40, 10% glycerol, 0.5% sodium deoxycholate, 1mM PMSF, 1 mM NaF, 1 mM Na3VO4 and 1% protease inhibitormixture (catalog no. P8340, Sigma). Cell lysates were placed on ice for 30 min to facilitate protein extraction, and then centrifuged at 12000 rpm for 30 min at 4 °C. The lysates were quantified by a BCA protein assay kit. Equal amounts proteins (30μg/lane) were loaded on a 10% or 12% Bis-Tris polyacrylamide gel for electrophoresis, and then were transferred onto a Nitrocellulose membrane (PALL, BioTrace, Mexico) at 4 °C. The membranes were blocked with 5% skim milk followed by incubation with appropriate primary antibody at 4 °C overnight, washed 7-min with PBST for four times and then probed with secondary antibody conjugated with horseradish peroxidase. The signals were visualized with Pierce ECL Western Blotting Substrate (Thermo Scientific, USA) and analyzed using the Image J software (USA). The antibodies used for western blotting are listed as follow: rabbit anti-UHRF2 polyclonal antibodies (abcam, ab28673), rabbit anti-DNMT1 antibodies (Active Motif, 39905), Rabbit anti-5hMC antibodies (Active Motif, 39791), Rabbit anti-TET2 antibodies (abcam, ab94580) and Rabbit anti-GAPDH (abcam, ab181602).
Two NSCLC cell lines, 60 NSCLC samples, and their adjacent nontumorous liver tissues were performed to extract RNA. Total RNA was isolated using Trizol reagent (Invitrogen), treated with TURBO DNase (Ambion,Life Technologies) and reverse transcripted using PrimeScript™ RT reagent Kit (Takara) with random hexamer primers.
Quantitative real-time PCRs were performed using Syber Green PCR Fast PCR Master Mix 2x (Applied Biosystems). The housekeeping genes β-actin were used for normalization of qRT-PCR data, unless otherwise stated. The qPCR primers used in this research were listed in Table 1.
Lists of primers sequence analyzed by Quantitative Real-time PCR
| UHRF2 | Forward | TTGCTGCTGATGAAGACGTT |
|---|---|---|
| Reverse | TTCTGCATCAAACCAGAATCC | |
| TET1 | Forward | TCTTCCCCATGACCACATCT |
| Reverse | GAGGGAAAAGAAGCCCAAAG | |
| TET2 | Forward | ACGCTTGGAAGCAGGAGAT |
| Reverse | CACAAGGCTGCCCTCTAGTT | |
| TET3 | Forward | CCCACAAGGACCAGCATAAC |
| Reverse | CCATCTTGTACAGGGGGAGA | |
| β-actin | Forward | TCCCTGGAGAAGAGCTACG |
| Reverse | GTAGTTTCGTGGATGCCACA |
Cell viability, wound healing assays and matrigel invasion assays
For cell viability assay, Cell proliferation was detected using the Cell Counting Kit-8 (CCK8, YESEN, Shanghai, China) according to the manufacturer's instructions. For wound healing assay, cells were incubated in 6-well plates and covered 95% of the plated bottom after 24h, and wound lines were scratched by 200μl pipette tip. Then migrating cells were measured under a microscope at 0 and 48 hours. For cell invasion assays, cells were incubated in 24-well transwells which precoated with matrigel (Falcon354480; BD Biosciences, USA). A total number of 2 × 104 were resuspended in 200μl RPIM-1640 or DMEM without FBS and were placed upper the transwell, and 600μl RPIM-1640 with 10% FBS were added under the transwells. The transwells were incubated with 5% CO2 at 37°C for 48 hours, then cells were fixed in 4% paraformaldehyde and stained by crystal violet. Cells in 5 random fields (magnification, ×100) were counted and photographed. All results are mean ± SEM of 3 independent experiments.
Cell Cycle and Cell Apoptosis Analysis
Cell cycle distribution was analyzed by flow cytometry as follows. After puromycin screened, cells were harvested and washed with PBS, and then fixed in 70% ice-cold ethanol for about 24 h at -20 °C.After that fixed cells were washed gently with cold PBS twice, incubated with 0.5 μg/mL RNase A for 30 min at 37 °C, and then stained with 50 μg/mL propidium iodide in the dark at room temperature for 30 min. The stained cells were detected by LSRFortessa Flow Cytometer (BD Biosciences) and calculated using FlowJo software. Apoptotic cells were evaluated in vitro by annexin-V-fluo- rescein isothiocyanate (PE) and 7-Amino-Actinomycin (7-AAD) using Annexin-V (Ruo) Apoptosis Detection Kit I (BD Pharmingen, San Jose, Calif) according to the manufacturer's protocol. Stained cells were then analyzed with LSRFortessa Flow Cytometer (BD Biosciences), and the data were analyzed using FlowJo software (Tree Star Inc., Ashland, Ore).
Statistics
Data are presented as the mean±SEM of replicate experiments (≥3). Results were analyzed using the SPSS 16.0 software (SPSS) and PRISM 5.0 (GraphPad Software Inc.). The Student unpaired t test or unpaired t test with Welch's correction was used to analyze intergroup differences for two groups, ANOVA was used to analyze more than two groups, and Pearson's correlation coefficient was used to analyze the correlation between groups. The cumulative survival time was calculated using the Kaplan-Meier method and analyzed using the log-rank test. Univariate and multivariate analyses were based on the Cox proportional hazards regression model. P values < 0.05 were considered statistically significant.
Result
UHRF2 was decreased in NSCLC tissues
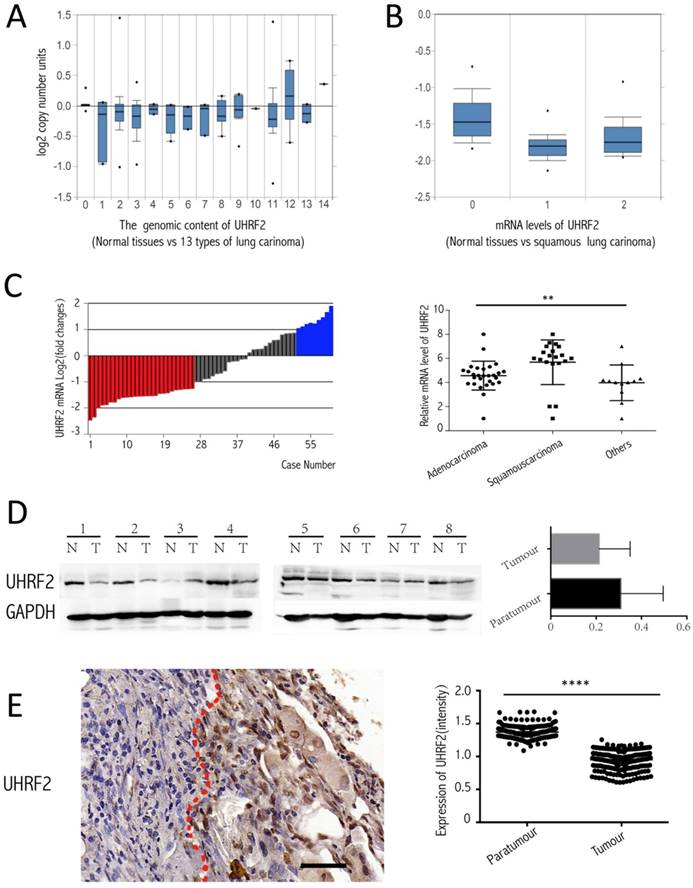
To investigate the expression of UHRF2 in NSCLC tissues compared with corresponding paratumorous tissues. Firstly, we analyzed the level of UHRF2 from the data of the Oncomine website (https://www.oncomine.org/resource/login.html), there are two corresponding research, and one is from TCGA lung 2 research (UCSC refGene, July 2009, hg18, NCBI 36.1, March 2006) which evaluated the copy number of UHRF2 in 1537 samples, another is from Talbot lung research [15] which evaluated UHRF2 in mRNA level from 93 samples. The data showed that UHRF2's copy number was decreased in majority of types of NSCLC (Figure 1A), also UHRF2 was dowregulated in mRNA level in squamous lung carcinoma (Figure 1B). Then, we performed RT-qPCR to assess UHRF2 expression in NSCLC tissues and paired adjacent non-tumor tissues from 60 NSCLC patients. We found that UHRF2 was downregulated in 43.33% of NSCLC specimens and upregulated in 15% specimens (Figure 1C), in terms of pathologic types, UHRF2 expressed lower in adenocarcinoma than that in squamous carcinoma or other types of lung cancer (Figure 1C). Furthermore,we use immunoblot assays to detect UHRF2 in 8 NSCLC tumour tissues and paired normal lung tissues.The results also indicated that UHRF2 was downregulated in NSCLC than that in corresponding para-tumorous lung tissues (Figure 1D). Finally, we employed the immunohistochemistry (IHC) to detect UHRF2 expression in tissues microarray containing 208 NSCLC and corresponding para-tumorous normal tissues. The results showed that UHRF2 was downregulated in NSCLC tumor tissue compared with the corresponding normal tissues (Figure 1E).
Knockdown UHRF2 promoted NSCLC proliferation and enhanced the invasion and migration of NSCLC cells
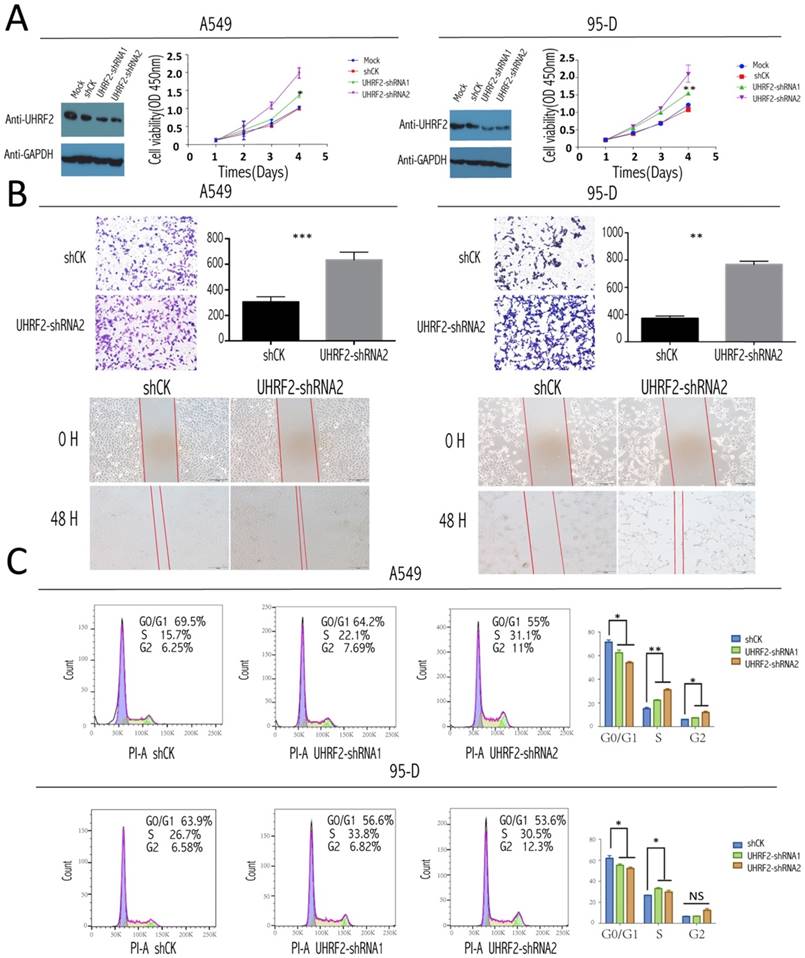
To test the role of UHRF2 in the tumorigenesis and progression of NSCLC, a loss of function assay was performed by infecting the NSCLC cell line: A549 and 95-D cells with lentivirus containing shRNAs targeting UHRF2 and lentivirus mediates shCK inserted plko vector as control group. After UHRF2 was effectively knocked-down determined by western blotting (Figure 2A), the result of CCK8 showed the knockdown of UHRF2 elevated the ability of cell proliferation compared with control group (Figure 2A). Transwell and scratch assays showed that UHRF2 knockdown cells was much enhanced in invasion and migration after lentivirus infected in 48hr (Figure 2B).
Role of UHRF2 in cell cycle of NSCLC cells
For the analysis of cell cycle, we stained this two cell lines with PI according to the protocol and analyzed them with FACS. The results showed that knockdown UHRF2 expression significantly decreased the percentage of cells in G1 phase, while increased the percentage of cells in G2 phase,and the percentage of cells in S phase also showed increased trend (Figure 2C). This increased trend became more obviously when combining S and G2 compared with G1and G0. Meanwhile, we didn't observe obvious difference when overexpressed UHRF2 in two cell lines. The result above was similar with the previous report [16].
UHRF2 is downregulated in NSCLC tissues and positively correlated with 5hmC. A. Oncomine database showed UHRF2 DNA content was downregulated in different types of lung carcinoma ( A indicated as follows: 0-No value,1-Acinar lung adenocarcinoma, 2-Lung adenocarcinoma, 3-Lung adenocarcinoma/mixed subtype, 4-Lung clear cell adenocarcinoma, 5-Lung mucinous adenocarcinoma, 6-Micropapillary lung adenocarcinoma,7-Mucinous cronchioloalveolar carcinoma,8- Non-mucinous bronchioloalveolar carcinoma, 9-Papillary lung adenocarcinoma,10-Solid lung adenocarcinoma,11-Squamous cell lung carcinoma,12-Squamous cell lung carcinoma/basaloid variant,13-Squamous cell lung carcinoma/papillary variant, 14-Squamous cell lung carcinoma/small cell variant). B. Oncomine database showed UHRF2 mRNA level was downregulated in squamous carcinoma (B indicated as follows: 0-No value, 1-Squamous cell lung carcinoma, 2-Tongue squamous cell carcinoma). C. UHRF2 levels were determined by RT-qPCR and normalized to β-actin in 60 paired NSCLC tumour and matched adjacent non-tumor tissues. Data were expressed as the log2 fold change (ΔCt [NSCLC/Non.]), and significant UHRF2 change was defined as a log2 fold change >1. The scatter plot diagram illustrated that the UHRF2 level in different histologic types of NSCLC tissues. D. Immunoblot analyzed the expression of UHRF2 in 8 patients (N: Normal tissues, T: tumour). The right bar plots showed the intensity(presented relative to those of Gapdh) of immunoblot assay. E. Immnohistochemistry in Tissue microarray of 208 NSCLC patients showed UHRF2 was loss in Tumour tissue compared with adjacent normal tissues and the relationship between them. Data are presented as means ± SEM. N.S. non-significant, P≥0.05; *P<0.05; ****P<0.0001; Scale bar 100 μm.
UHRF2 knockdown enhanced NSCLC cell proliferation, invasion,and desregulated cell cycle. A. The efficacy of influence of UHRF2 was determined by western blotting (WB) in A549 and 95-D cell respectively (GAPDH as an internal control). The coloured scatter plot illustrated the Cell viability in four different record time points determined by the absorbance at OD 450nm two h after CCK8 was added. B. Transwell assays showed knockdown UHRF2 in A549 and 95-D cell lines enhanced the ability of migration and invasion and the bar plot showed the data converted into statictics(Upper half part).Wound healing assays of A549 and 95-D with gene UHRF2 interfered(compared to control group) were photographed in 0 hour and 48 hour after wound cells. C. Cell cycle analysis of shCK, PLKO-shRNA1 and PLKO-shRNA2 knockdown A549 and 95-D cells by flow cytometry, the top right number showing the percentage of G0/G1, S or G2 phase of cells.the bar polts on the right of each row illustrated the change of cell cycle phase much clearer. Data were presented as means ± SEM. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001; NS: no significant. Data were representative of more than three independent experiments.
Elevated UHRF2 Expression Induced Apoptosis of NSCLC Cells and Abated the Invasion, Proliferation and Migration of NSCLC Cells
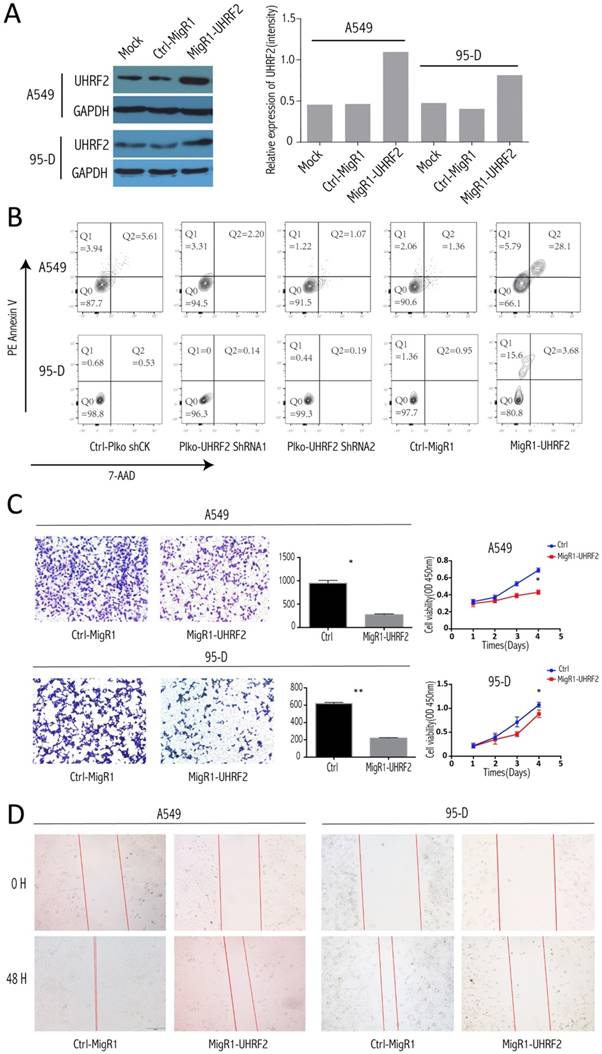
To explore whether UHRF2 can influence apoptosis of NSCLC cell, we conducted assay in different treated cell group (UHRF2-downregulated, UHRF2 overexpression and control groups). After elevated the UHRF2 expression well (Figure 3A), the apoptosis was determined by FACS, the results showed that overexpressing UHRF2 in A549 and 95-D cells can significantly induce cell apoptosis (Figure 3B). The percentage of Q1 (early apoptosis) + Q2 (late apoptosis) of cells was significantly higher than others. Additionally, we founded that the elevated UHRF2 expression inhibited cell migration, invasion and proliferation of A549 and 95-D cells (Figure 3C and 3D).
UHRF2 possibly Excerted Influence on TET family Enzymes, Especially TET2 and UHRF2 was closely Associated with Genomic Content of 5-hmC
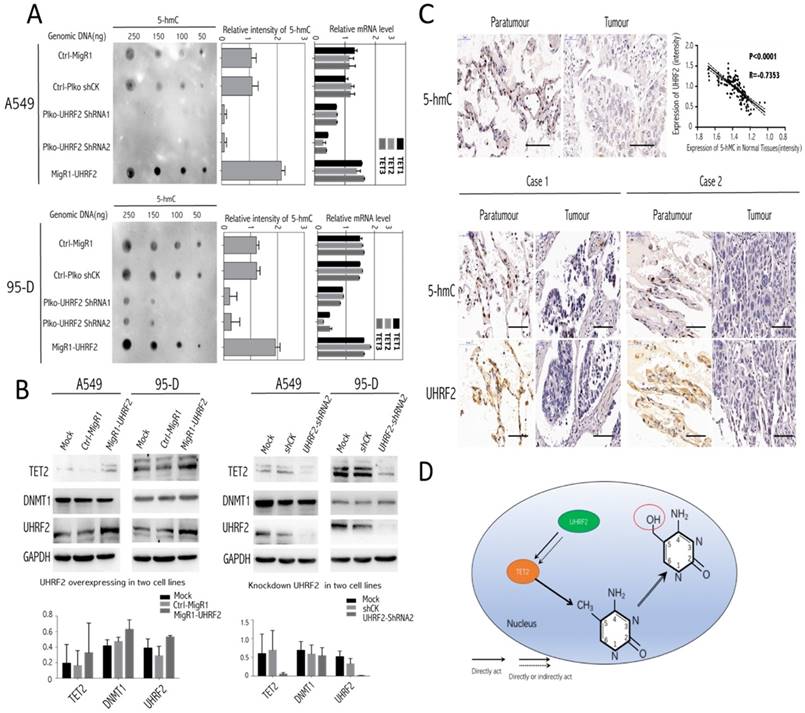
Here, we tried to investigate the underlying mechanism involving it. The present evidence showed that UHRF2 was possibly correlated with 5-hmC content [13], thus we conducted Dot-blot assay to examine 5-hmC level in different genomic DNA content extracted from A549 and 95-D. The results indicated that the 5-hmC level was positively associated with UHRF2 expression (Figure 4A). Moreover, the level of 5-hmC was diminished in cells with UHRF2 knockdown, while was increased in cells overexpressing UHRF2.
TET enzymes oxidize 5-methylcytosine (5-mC) to 5-hydroxymethylcytosine (5-hmC), which can lead to DNA demethylation [17]. So, we analyzed TET family (TET1, TET2, TET3) mRNA level in A549 and 95-D cells transfected with UHRF2 interference or overexpression lentivirus. We found that mRNA level of TET genes was significantly decreased in UHRF2 knockdown cells and increased in UHRF2 overexpressed cells (Figure 4A). Furthermore, we test DNMT1, TET2 by immunoblotting in cell lines which were interfered with UHRF2 expression. The results indicated that TET2 would be affected by UHRF2 expression (Figure 4B). TET2 expression was positively associated with UHRF2 expression (Figure 4B). Additionally, DNMT1 also showed slightly change. we also evaluated the UHRF2 expression and 5-hmC level in the NSCLC and corresponding normal tissues, and we founded that the level of 5-hmC is lower in NSCLC tissues than that in corresponding normal tissues, which in line with the IHC results of UHRF2. In NSCLC tissues, the expression of UHRF2 is positively corrrelated with the level of 5-hmC (Figure 4C). Thus, we speculated that UHRF2 function as a tumor supressor by influenceing the level of 5-hmC (Figure 4D).
Low Expression of UHRF2 Correlated Significantly With the Poor Prognosis of NSCLC Patients
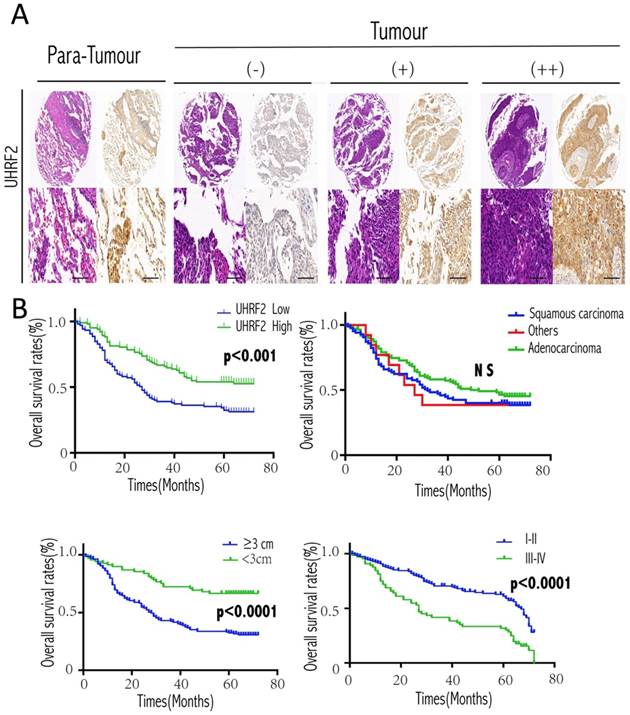
Then, we further analyzed the correlation between UHRF2 and clinicopathological characteristics in 208 NSCLC cases. Low expression of UHRF2 accounted for 50.96% specimens from patients with NSCLC (106 of 208 patients). The correlations between the expression of UHRF2 and clinicopathological features were shown in Table 2. We found that UHRF2 expression levels were correlated with tumor size (p<0.001), histological type (p=0.019), tumor differentiation (p=0.04) and age. However, other clinicopathological features, including gender, smoking status, lymph node metastasis were not directly associated with the expression of UHRF2. Notably, univariate analysis showed that Lymph node metastasis, tumor size, tumor stage, UHRF2 level were associated with OS (overall survival). Moreover, multivariate analysis showed lymph node metastasis, tumor size and UHRF2 were related to OS (Table 3). Univariate and multivariate investigation revealed that UHRF2 was an independent risk marker of the OS. The level of UHRF2 protein was founded to be incrediblly heterogeneous in NSCLC samples, and representative images were shown in Figure 5A, UHRF2 showed high expression in normal tissues whereas various level of UHRF2 was detected in tumor tissues (-, absent; +, weak; ++, strong).
At the end of follow-up, 120 patients had died, and the 5-year OS rate was 42%. The 5-year overall survival rate for patients with low UHRF2 expression was significant lower than patients with high UHRF2 expression (p<0.001, Figure 5A). As our qPCR results of NSCLC tissues showed the level of UHRF2 expression was different between squamous cell carcinoma and adenocarcinoma, we performed a subgroup analysis by pathological subtype, but the results was no significant (Figure 5B). In addition, patients with a larger tumor size and poorer differentiation stage possessed a more unfavorable OS (Figure 5B).
Overexpression of UHRF2 induced NSCLC cell apoptosis and abated the capability of invasion and migration. A. The efficiency of migR1-UHRF2 overexpression was determined by immunoblotting (left panel). Bar plot present the efficient of UHRF2 overexpression in quantitative value(right panel). B. Overexpressing UHRF2 in A549 and 95-D can induce cell apoptosis,but downregulating UHRF2 didn't show different compared with control group. C. The left panel showed the result of Transwell assays, bar plots in the middle part indicated the definite cell numbers of the transwell assays.CCK8 assays(right panel) indicated that overexpressing UHRF2 in A549 and 95-D would abate the capability of invasion and proliferation. D. Overexpressing UHRF2 could slow down the migration of A549 and 95-D cell lines. Data are representative of more than three independent experiments.
Genomic 5-hmC level was positively correlated with UHRF2 content via regulating TET2. A. The dot blot results showed the expression of UHRF2 was correlated with 5-hmC content in different dose of DNA (250ng, 150ng, 100ng, 50ng). The bar plots in the middle showed the quantitative value of dot blot intensity.the right bar plots showed the qPCR results of each cell group in TET family enzymes. B. A549, 95-D was performed to identify the influence of UHRF2 on DNMT1 and TET2 by immunoblotting assays and bar graph indicated overexpression or knockdown of UHRF2 and the corresponding DNMT1 and TET2 level. C. Immunohistochemistry of 5-hmC and UHRF2 in tumour and paratumour normal tissues. The lower part was illustrated with the expression of UHRF2 and 5-hmC of two typical NSCLC patients. D. Diagram of speculating the interactive correlation between UHRF2 and 5-hmC. Data are presented as means ± SEM. Data are representative of more than three independent experiments. Scale bar 100 μm.
Correlation between UHRF2 and clinicopathological characteristics in 208 NSCLCs.
| Variables | No. of patients | UHRF2 expression level | ||
|---|---|---|---|---|
| low | high | P | ||
| Age | ||||
| <60 | 102 | 42 | 60 | 0.008 |
| ≥60 | 106 | 64 | 42 | |
| Gender | ||||
| Male | 148 | 75 | 73 | 1.000 |
| Female | 60 | 31 | 29 | |
| Smoking status | ||||
| Smokers | 84 | 39 | 45 | 0.323 |
| Non-smokers | 124 | 67 | 57 | |
| Histological type | ||||
| Squamous cell carcinoma | 85 | 37 | 48 | |
| Adenocarcinomas | 110 | 58 | 52 | 0.019 |
| Othera | 13 | 11 | 2 | |
| Tumor stage | ||||
| I-II | 144 | 67 | 77 | 0.071 |
| III-IV | 64 | 39 | 25 | |
| Lymph node metastasis | ||||
| Yes | 90 | 48 | 42 | 0.578 |
| No | 118 | 58 | 60 | |
| Tumor size | ||||
| <3 cm | 69 | 32 | 37 | <0.001 |
| ≥3 cm | 139 | 100 | 39 | |
| Differentiation | ||||
| Well/moderate | 115 | 30 | 85 | 0.04 |
| Poor | 93 | 71 | 22 | |
NOTE: Bold values are statistically significant (P< 0.05).
a Other including adenosquamous carcinoma, large-cell carcinoma, mucoepidermoid carcinoma and carcinosarcoma. * P value was analyzed by squamous cell carcinomas vs. adenocarcinomas.
Expression of UHRF2 and prognostic significance in NSCLC patients. A. Expression of UHRF2 in 208 cases of NSCLC tissues and matched non-tumor tissues was detected by immunohistochemistry. The IHC picture magnified in 40× (upper) and 200×(lower). B. NSCLC patients expressing low levels of UHRF2 had the poorest prognosis in terms of OS. Different histologic types; tumor size and disease stage were also assessed with the overall survival. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001; NS no significant. Scale bar 100 μm.
Univariate and multivariate analysis of factors associated with OS.
| Variables | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | p | HR | 95% CI | p | ||
| Gender | |||||||
| (male vs. female) | 0.789 | 0.526-1.183 | 0.251 | ||||
| Smoking status | |||||||
| (non-smokers vs. smokers) | 0.779 | 0.543-1.118 | 0.175 | ||||
| Differentiation | |||||||
| (well/moderate vs. poor) | 1.431 | 1.000-2.049 | 0.050 | ||||
| Lymph node metastasis | |||||||
| (yes vs. no) | 3.042 | 2.103-4.399 | <0.001 | 2.415 | 1.537-3.793 | <0.001 | |
| Tumor size | |||||||
| (≥3cm vs.<3cm) | 2.755 | 1.758-4.318 | <0.001 | 2.101 | 1.312-3.363 | 0.002 | |
| Tumor stage | |||||||
| (III-IV vs. I-II) | 2.771 | 1.922-3.993 | <0.001 | 1.423 | 0.909-2.226 | 0.123 | |
| UHRF2 level | |||||||
| (high vs. low) | 1.906 | 1.321-2.750 | =0.001 | 2.048 | 1.403-2.990 | <0.001 | |
Abbreviations and note: OS, overall survival; 95% CI, 95% confidence interval; multivariate analysis, Cox proportional hazards regression model. Variables were adopted for their prognostic significance by univariate analysis with forward stepwise selection (forward, likelihood ratio). Variables were adopted for their prognostic significance by univariate analysis (p < 0.05).
Discussion
Cancers subvert both the genome and the epigenome to evolve mechanisms by which tumour cells can escape growth control and surveillance to become increasingly autonomous of the requirements of the host. The involvement of altered chromatin in cancer has been such apparent since the early days of pathology diagnosis through light microscopic observations. The 5-hmC epigenetic mark was first identified in the T-even bacteriophage almost six decades ago [18]. 5-hmC is considered to be an oxidized 5-mC derivatives (mainly including 5-hmC, 5-fC and 5-caC). It is clear that these oxidized 5-mC derivatives serve as DNA demethylation intermediates that are important for programming and reprogramming during development and differentiation of cell [19]. So far, many studies provided solid evidence that 5-hmC commonly reduced in multiple cancer [8, 20-22]. Our study determined that 5-hmC was significantly decreased in NSCLC tumour tissues compared with adjacent normal tissues, which coincides with previous studies. Mechanistically, in embryonic stem cells all genome-wide maps of 5-hmC in human ESCs and mouse ESCs indicate that 5-hmC tends to exist in gene bodies, promoters, and enhancers [23]. So 5-hmC appears to function as a regulator of the gene transcriptional activity [19, 24].
Most evidences have shown that 5-hmC interact with some molecular readers reciprocally during demethylation process, such as UHRF2 [13, 25]. UHRF2 generally is deemed to be a nuclear E3 ubiquitin ligase which is involved in cell cycle and epigenetic regulation. UHRF2 was reported to interact with many key factors in cell cycle [4, 7, 26]. Our result indicated that loss of UHRF2 can promote NSCLC cell entering S or G2 phase. Knockdown UHRF2 enhanced the proliferation, invasion and migration of NSCLC cell. This result is accord with some studies which indicate UHRF2 probably is a tumor suppressor [7, 26, 27]. Importantly, UHRF2 was reported to be a transcriptional target of E2F1 by directly interaction, and was required for E2F1 induction of apoptosis and transcription of a number of important apoptotic regulators [28]. Our study revealed that overexpressing UHRF2 in NSCLC cells could induce cell apoptosis, which further highlight that UHRF2 tend to be a tumor suppressor.
To further investigate the probable mechanism underlying UFRF2 function. We performed dot-blot to detect the correlation between UHRF2 and 5-hmC, we found the DNA 5-hmC level was positively correlated with UHRF2 level. Knockdown or overexpressing UHRF2 in NSCLC cells could down- or up-regulate 5-hmC level simultaneously.
It is well accepted that dynamic DNA methylation and demethylation mainly catalyzed by two kinds of protein, DNMT and TET family. For example, 5-hmC is generated from 5-mC oxidized by TET family [29]. The transformative discovery that TET family enzymes can oxidize 5-mC has greatly advanced our understanding of DNA demethylation. 5-hmC is a key nexus in demethylation that can either be passively depleted through DNA replication or actively reverted to cytosine through iterative oxidation and thymine DNA glycosylase (TDG)-mediated base excision repair[17, 30, 31]. Among TET protein family, TET2 is an important member which mutations of TET2 are frequently observed in malignant tumorgenesis so that influence the tumor development [32, 33].Thus, we detected the DNMT1 and TET2 expression after UHRF2 interference. We found DNMT1 is not interfered by UHRF2 level, but TET2 was obviously different during interfering UHRF2 expression. That inspired us the probable mechanism underlying the correlation between UHRF2 and 5-hmC. Therefore,we suggest an attractive model which UHRF2 probably affect generating 5-hmC via TET2 directly or indirectly, consequently maintaining the cell normality.
As some study has reported that key factors in the 5-hmC generating pathway can be a therapeutically targeted to restore 5-hmC in human [20], thus our findings could reveal new strategies for the design of cancer treatment via regulating epigenome modification.
Acknowledgements
This study was funded by the (320.6799.15055) WU JIEPING medical foundation and Talent funding programme of Zhongshan affiliated hospital to Fudan University.
Abbreviations
NSCLC, Non-small cell lung carcinoma; UHRF2, ubiquitin like with PHD and ring finger domains 2; 5-hmC, 5-methylcytosine hydroxylation, TET2, ten-eleven translocation 2.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7-30
2. Hirsch FR, Suda K, Wiens J, Bunn PA Jr. New and emerging targeted treatments in advanced non-small-cell lung cancer. Lancet. 2016;388:1012-24
3. Zhou T, Xiong J, Wang M, Yang N, Wong J, Zhu B. et al. Structural Basis for Hydroxymethylcytosine Recognition by the SRA Domain of UHRF2. Molecular Cell. 2014;54:879-86
4. Chen W, Wu M, Hang T, Wang C, Zhang X, Zang J. Structure insights into the molecular mechanism of the interaction between UHRF2 and PCNA. Biochem Biophys Res Commun. 2017;494:575-80
5. Lu S, Yan D, Wu Z, Jiang T, Chen J, Yuan L. et al. Ubiquitin-like with PHD and ring finger domains 2 is a predictor of survival and a potential therapeutic target in colon cancer. Oncol Rep. 2014;31:1802-10
6. Lu H, Bhoopatiraju S, Wang H, Schmitz NP, Wang X, Freeman MJ. et al. Loss of UHRF2 expression is associated with human neoplasia, promoter hypermethylation, decreased 5-hydroxymethylcytosine, and high proliferative activity. Oncotarget. 2016;7:76047-61
7. Mori T, Ikeda DD, Fukushima T, Takenoshita S, Kochi H. NIRF constitutes a nodal point in the cell cycle network and is a candidate tumor suppressor. Cell Cycle. 2011;10:3284-99
8. Li X, Liu Y, Salz T, Hansen KD, Feinberg A. Whole-genome analysis of the methylome and hydroxymethylome in normal and malignant lung and liver. Genome Res. 2016;26:1730-41
9. Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89-97
10. Song CX, Yi C, He C. Mapping recently identified nucleotide variants in the genome and transcriptome. Nat Biotechnol. 2012;30:1107-16
11. Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA. et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300-3
12. Chen K, Zhang J, Guo Z, Ma Q, Xu Z, Zhou Y. et al. Loss of 5-hydroxymethylcytosine is linked to gene body hypermethylation in kidney cancer. Cell Res. 2016;26:103-18
13. Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C. et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146-59
14. Liao YF, Wu YB, Long X, Zhu SQ, Jin C, Xu JJ. et al. High level of BRD4 promotes non-small cell lung cancer progression. Oncotarget. 2016;7:9491-500
15. Talbot SG, Estilo C, Maghami E, Sarkaria IS, Pham DK, P Oc. et al. Gene expression profiling allows distinction between primary and metastatic squamous cell carcinomas in the lung. Cancer Res. 2005;65:3063-71
16. Wu TF, Zhang W, Su ZP, Chen SS, Chen GL, Wei YX. et al. UHRF2 mRNA expression is low in malignant glioma but silencing inhibits the growth of U251 glioma cells in vitro. Asian Pac J Cancer Prev. 2012;13:5137-42
17. He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303-7
18. Wyatt GR, Cohen SS. The bases of the nucleic acids of some bacterial and animal viruses: the occurrence of 5-hydroxymethylcytosine. Biochem J. 1953;55:774-82
19. Song CX, He C. Potential functional roles of DNA demethylation intermediates. Trends Biochem Sci. 2013;38:480-4
20. Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K. et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150:1135-46
21. Song CX, Yin S, Ma L, Wheeler A, Chen Y, Zhang Y. et al. 5-Hydroxymethylcytosine signatures in cell-free DNA provide information about tumor types and stages. Cell Res. 2017;27:1231-42
22. Kudo Y, Tateishi K, Yamamoto K, Yamamoto S, Asaoka Y, Ijichi H. et al. Loss of 5-hydroxymethylcytosine is accompanied with malignant cellular transformation. Cancer Sci. 2012;103:670-6
23. Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A. et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368-80
24. Shen L, Wu H, Diep D, Yamaguchi S, D'Alessio AC, Fung HL. et al. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153:692-706
25. Zhou T, Xiong J, Wang M, Yang N, Wong J, Zhu B. et al. Structural basis for hydroxymethylcytosine recognition by the SRA domain of UHRF2. Mol Cell. 2014;54:879-86
26. Mori T, Ikeda DD, Yamaguchi Y, Unoki M, Project N. NIRF/UHRF2 occupies a central position in the cell cycle network and allows coupling with the epigenetic landscape. FEBS Lett. 2012;586:1570-83
27. Bai L, Wang X, Jin F, Yang Y, Qian G, Duan C. UHRF2, another E3 ubiquitin ligase for p53. Biochem Biophys Res Commun. 2012;425:908-11
28. Lu H, Hallstrom TC. The nuclear protein UHRF2 is a direct target of the transcription factor E2F1 in the induction of apoptosis. J Biol Chem. 2013;288:23833-43
29. Koivunen P, Laukka T. The TET enzymes. Cell Mol Life Sci. 2017
30. Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472-9
31. Yang J, Guo R, Wang H, Ye X, Zhou Z, Dan J. et al. Tet Enzymes Regulate Telomere Maintenance and Chromosomal Stability of Mouse ESCs. Cell Rep. 2016;15:1809-21
32. Hu L, Li Z, Cheng J, Rao Q, Gong W, Liu M. et al. Crystal structure of TET2-DNA complex: insight into TET-mediated 5mC oxidation. Cell. 2013;155:1545-55
33. Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A. et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553-67
Author contact
![]() Corresponding author: Jian-Yong Ding, M.D., Ph.D., Department of Thoracic Surgery, The Affiliated Zhongshan Hospital of Fudan University, 180 Fenglin Road, Shanghai, P.R. China, 200032 Tel. & Fax: +86-21-64041990 E-mail: dingjianyongmdcom
Corresponding author: Jian-Yong Ding, M.D., Ph.D., Department of Thoracic Surgery, The Affiliated Zhongshan Hospital of Fudan University, 180 Fenglin Road, Shanghai, P.R. China, 200032 Tel. & Fax: +86-21-64041990 E-mail: dingjianyongmdcom

 Global reach, higher impact
Global reach, higher impact