3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(11):2046-2053. doi:10.7150/jca.24474 This issue Cite
Research Paper
A Study on Effect of Oxaliplatin in MicroRNA Expression in Human Colon Cancer
1. School of Medical Sciences, Örebro University, SE 70182 Örebro, Sweden.
2. Department of Oncology and Department of Clinical and Experimental Medicine, SE-581 83, Linköping University, Linköping, Sweden.
3. Faculty of Allied Health Sciences, Chettinad Academy of Research and Education, Kelambakkam, India, 603 103.
# Contributed equally to the work.
Received 2017-12-21; Accepted 2018-3-14; Published 2018-5-22
Abstract
Colorectal cancer is a commonly diagnosed malignancy and also the major cause of death worldwide. Chemotherapy is the primary therapy for advanced colorectal cancer. Although oxaliplatin has potential effect in colorectal cancer therapy, the molecular mechanisms involved in its cytotoxic effects are not well elucidated. This study outlines the regulatory effects of oxaliplatin on miRNAs expression in colon cancer cells and correlates it with the changing microRNA expression with p53 and p73 expression status in cells. HCT116p53+/+ and HCT116p53-/- cells were exposed to oxaliplatin, and the cellular viability was determined by XTT. p73 was knocked down using siRNA and the tumor cells were then treated with oxaliplatin. The expression profile of 384 miRNAs was determined by TaqMan® human miRNA array and calculated by the ∆∆Ct method. Cellular viability was found to decrease after the treatment with oxaliplatin in a dose-dependent manner. The wild-type p53 cells were found to be more sensitive than the null-p53 derivatives. A selective set of miRNAs were either up-regulated or down-regulated in response to the oxaliplatin treatment with a presumable role of p53 and p73 proteins. The miRNAs expression is known to influence the pharmacodynamic mechanisms of oxaliplatin and these effects have been observed to be regulated by p53 and p73. Our results may therefore provide more evidence for identifying a suitable biomarker for the diagnosis of colon cancer.
Keywords: Oxaliplatin, miRNAs, p53, p73, colon cancer cells.
Introduction
Colorectal cancer is statistically reported to be one among the most commonly diagnosed cancers, catastrophically affecting more than 1.4 million people with a high death rate every year of about 693,900 deaths worldwide [1]. Chemotherapy is a universally acclaimed effective primary treatment in both resectable and advanced colorectal cancer conditions which involves anti-cancer drugs or compounds to kill the cancer cells. Studies proved that Oxaliplatin, a compound from third generation diaminocyclohexane platinum family showed positive effects in the treatment of colorectal cancer [2]. It belongs to the family of platinum analogous to carboplatin and cisplatin and has been known for several years for its chemotherapeutic compounds [3]. Despite the widespread clinical use of oxaliplatin, the molecular mechanisms behind its cytotoxic effects have not been completely understood. Further, clinical prognostic markers of response to oxaliplatin remain poorly characterized.
MicroRNAs commonly termed as miRNAs belong to a group of small non-coding RNAs of 18-24 nucleotides which are known to significantly regulate gene expression, post translational repression or cleavage of the mRNA targets [4]. The most phenomenal property of miRNAs is its ability to regulate various biological processes such as cell proliferation, differentiation and apoptosis [5]. Remarkably miRNAs have be known to be involved in various cancer associated pathways which is emphasized by the fact that half of all miRNA genes are in cancer-associated genomic regions or fragile sites [6] and the expression of these miRNA genes have been observed to be dysfunctional in cancer tissues when compared to normal tissue, including colorectal cancer [7]. Most miRNAs behave as oncogenes and tumor suppressor genes depending upon the target mRNA and play a critical role in the pathogenicity of various malignancies including colon cancer [8]. Further, a collective amount of evidence suggests the significant function of miRNAs in modulating chemo-sensitivity and chemo-resistance of tumor cells [9]. When compared to other cancer types, dysregulation of miRNA expression in colon cancer has been scrutinized less extensively. But several miRNAs with potent clinical and biological relevance have been identified to have a link to various biological processes that are noteworthy for tumor progression and chemotherapy response [10]. A substantial proportion of target genes including oncogenes and anti-oncogenes have been regulated by specific miRNA. Many miRNAs influence the cellular response to anticancer agents by regulating several target mRNA through various events such as DNA repair systems, survival and cell death pathways, specific drug targets, or drug transport and metabolism.
p53, a cellular tu.mor suppressor protein plays an active role in the cellular response to carcinogenic agents. DNA-damaging compounds, which are p53-mutated have been found to be resistant to drug-induced apoptosis [11]. When this gene receives a signal through cellular stresses like DNA damage, it is found to activate the transcription of genes that are responsible for apoptosis process [12]. Studies revealed that cancerous cells in the colon with wild-type p53 resulted in cell cycle arrest and apoptosis when compared to oxaliplatin treated cells [13]. p73, which is a homologue to p53, is also activated by a variety of DNA damaging drugs. This p73 gene when silenced by siRNA resulted in evident resistance to chemotherapeutic drugs [14], including oxaliplatin [15]. p73 also activates the transcription of p53 responsive genes since it has homologous character like p53 and instigates apoptosis in p53 based manner [16]. Although p53 significantly functions as a transcription factor in regulating miRNA expression, [17] the role of p73 in this context is less investigated.
In this study, we hypothesize that the pharmacodynamic mechanisms of oxaliplatin may depend on its miRNAs expression. Therefore, we investigate the plausible outcome following exposure to oxaliplatin and its impact on the modulated expression of miRNAs in colon cancer cells, and besides that prove the involvement of p53 and p73 status in regulating the expression levels of miRNAs.
Materials and Methods
Characteristic conditions of colon cancer cell lines
Human colon carcinoma cell lines HCT116p53+/+ and HCT116p53-/- were a kind gift from Dr. Vogelstein (Johns Hopkins University). HCT116p53+/+ has the ability to produce wild-type p53, while HCT116p53-/- is devoid of both alleles of p53 which is being eliminated by process of homologous recombination [18].
Experimental design and oxaliplatin cytotoxicity assay
Cellular viability was measured using the Cell Proliferation Kit II (XTT) and the colon cancer cells were seeded at a density of 15,000 cells/cm2 in 96 well plates and then incubated approximately for 24 hours to allow them to attach and proliferate. After 24 hours, freshly prepared oxaliplatin (Sigma Aldrich, Stockholm, Sweden) was supplemented with a concentration ranging from 0-60 µM. After about 48 hours, the cells treated with oxaliplatin were subjected to cellular viability determination using the XTT assay, and the number of viable cells after the treatment was determined by measuring absorbance at 450 nm.
siRNA transfection
Knock down gene expression of the p73 isoform TAp73 was performed using 21bp of double-stranded oligonucleotide RNAs, forming a 19 bp duplex core with 2 nucleotides 3′ overhang.
In order to confirm the absence of similar homology to any additional known coding sequences of human genome, BLAST a highly exploited pairwise alignment tool was used to analyze the specifically selected siRNA sequences. Lipofectamine® RNAiMAX (Invitrogen) was utilized to perform the reverse transfection of colon cancer cells implementing siRNA of about 20nm concentration. Prior to administration of oxaliplatin, the cells were allowed to recover within a period of 24 hours.
Oxaliplatin treatment of colon cancer cells
The cancerous cells were suspended at a density of 25,000 cells/cm2 in 6- or 12-well plates. After 24 hours, freshly prepared oxaliplatin (Sigma Aldrich) of 2 µM concentration was added. The cells were further cultured under the conditions described above for approximately 48 hours prior to harvesting cells for protein and RNA extraction.
Protein expression assay
RIPA buffer containing 1% Protease Inhibitor Cocktail (Sigma-Aldrich) was employed to lyse the cells, after 48 hours of oxaliplatin treatment. The total protein concentration found in the cells was determined by the BCA Protein Assay (Pierce, Woburn, MA) after which the samples of about 20 µg protein were subjected to electrophoresis on precast Mini Protean TGX gels, 4-15% (Bio-Rad, Hercules, CA) and electro transferred to PDVF membranes (Bio-Rad). The membranes were incubated with mouse monoclonal antibody raised against TAp73 (1:500) (Abcam, Cambridge, MA) at 4ºC overnight, followed by the inclusion of a secondary HRP- conjugated goat-anti-mouse antibody (1:1000, DAKO Cytomation, Glostrup, Denmark) which was then incubated at room temperature for an hour. The proteins that adhered to the membranes were distinguished using the Amersham ECL plus Western Blot detection system including the specific secondary antibody against the primary monoclonal antibody (Amersham biosciences/GE Healthcare).
Extraction of RNA and reverse transcription
Total RNA was acquired from the cells after 48 hours of oxaliplatin treatment using the mirVana™ miRNA Isolation Kit (Invitrogen). TaqMan® MicroRNA reverse transcription kit and Megaplex™ RT primers, human pool A v2.0 (Applied Biosystems, Foster City, CA) was utilized to undertake the reverse transcription reaction. The obtained cDNA samples were pre-amplified using Megaplex™ PreAmp primers and TaqMan® Preamp master mix (Applied Biosystems).
Gene sequencing
| S. no | Gene name | Primers |
|---|---|---|
| 1 | TAp73 | Sense:5′-CGGAUUCCAGCAUGGACGU-3′ Antisense 5′-ACGUCCAUGCUGGAAUCCG-3′ |
| 2 | control non-silencing | sense:5′-UUCUCCGAACGUGUCACGU-3′; Anti-sense:5′-ACGUGACACGUUCGGAGAA-3′. |
Data analysis and statistics
For oxaliplatin cytotoxicity assay, the experiments were repeated thrice, and the cellular viability was expressed as mean values ± standard error deviations. The significance of the values obtained was analyzed using the statistical t-test. miRNA qPCR arrays, the expression levels of about 379 microRNAs and 5 control assays were analyzed using the TaqMan® MicroRNA Array Set v2.0, Card A (Applied Biosystems). Raw Ct-values (Cycle threshold) were calculated using the SDS software (Applied Biosystems). The R programming software was employed to normalize the raw Ct-values [19], along with qPCRNorm quantile normalization which is a data-driven normalization strategy for high-throughput qPCR data [20].
For the target prediction of miRNAs after the treatment of oxaliplatin, the miRNAs whose expression was altered more than 2-fold were selected. For the target prediction, two prediction programs, such as TargetScan Release 6.2 (http://www.targetscan.org) and MicroCosm Targets Version 5 (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/), were utilized and the target genes predicted by both the programs were identified and recorded.
Results
Effects of oxaliplatin on cellular viability
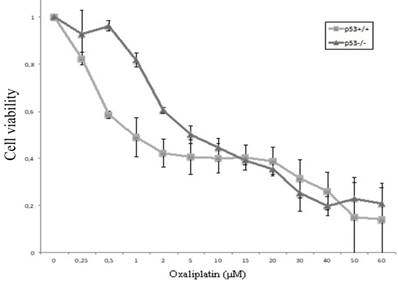
Cellular viability of HCT116 cells (HCT116p53+/+ and HCT116p53-/-) after exposure to oxaliplatin of increasing concentrations (0-60 μM) was assessed using the XTT assay. The IC50 value for HCT116p53+/+ cells and HCT116p53-/-cells was observed to be 1 μM, and 5 μM (Figure 1).
Chemosensitivity of HCT116p53+/+ and HCT116p53-/- colon cancer cells. After allowing the cells to attach in 96-well plates for 24 hours, oxaliplatin was added with a final concentration ranging from 0 - 60 µM. After 48 hours, cellular viability was determined using the XTT assay. Three independent experiments were performed in triplicate and the data represents the mean ± SD.
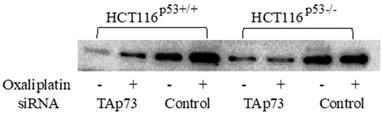
Protein expression of TAp73 after siRNA knockdown and oxaliplatin exposure
The TP73 gene was observed to give rise to different isoforms due to the modifications attained by the presence of alternative promoters as well as post transcriptional modifications like mRNA splicing. Previous research studies conducted related to TP73 have found that the full length TAp73 isoforms were considered to have tumor suppressor functions as they share many common target genes with p53 [21], while the truncated ΔNp73 isoforms were found to inhibit the function of TAp73 and p53 [22]. Protein expression of TAp73 after gene knockdown by siRNA and oxaliplatin treatment was analyzed using Western blot analysis. The doubled-stranded RNA oligonucleotides analogous to 5′-specific TAp73 sequences were found to be lacking in the ΔNp73 sequences that were used to precisely knock-down TAp73 expression in HCT116p53+/+, HCT116p53-/- cell lines [14]. After the transfection, the cells treated with oxaliplatin indicated a prominent decline in Tap73 expression when compared to non-interfering siRNA-transfected cells after 72 hours of transfection (Figure 2).
Expression of TAp73 examined by Western blot, Protein expression of TAp73 after gene knockdown by siRNA and oxaliplatin treatment in colon cancer cell lines HCT116p53+/+ and HCT116p53-/-. The cells were treated with oxaliplatin (2 μM) 24 hours after transfection. 72 hours post-transfection, TAp73 expression became markedly reduced compared to non-interfering siRNA-transfected cells.
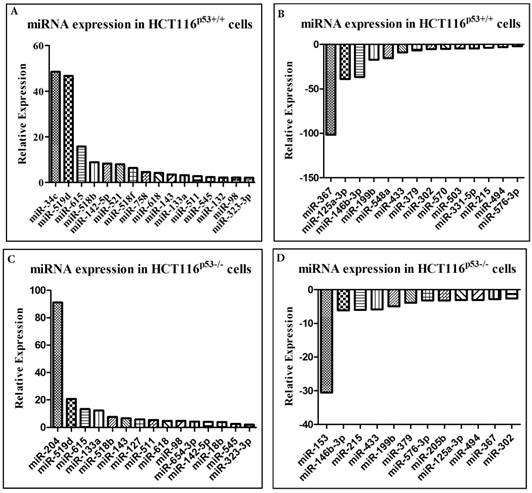
miRNA expression in colon cancer cells after oxaliplatin treatment
The manifestation of various genes of 379 miRNAs along with 5 control assays were analyzed in human colon cancer cell lines HCT116p53+/+ and HCT116p53-/- after oxaliplatin treatment. Around 210 and 267 of miRNAs from a total of 384 miRNAs were expressed in the samples. The data was expressed as N-fold change on comparing the oxaliplatin treated samples with untreated control. The oxaliplatin treatment impacted a diverse group of miRNAs and the outcome varied depending on the status of p53 and p73 genes. Particularly, those miRNAs which exhibited a two-fold adaptation between control and oxaliplatin treated cells were contemplated.
After exposure of oxaliplatin in the HCT116p53+/+ cell line, 16 miRNAs were found to be up-regulated (Figure 3a) and 14 miRNAs were noted to be down-regulated (Figure 3b) whereas in the HCT116p53-/- cells it was found that 15 miRNAs were highly elevated (Figure 3c) and 12 miRNAs were highly down-regulated (Figure 3d). It was observed that among the highly expressed miRNAs, 11 were common in both HCT116p53+/+ and HCT116p53-/- cell lines and within the down-regulated miRNAs, 10 were common to both the cell lines after the treatment. About 5 miRNAs were found to be elevated and 4 miRNAs were minimized exclusively in HCT116p53+/+ cell lines. Corresponding figures for HCT116p53-/- included 4 highly elevated miRNAs and 2 decreased miRNAs. Most of them were still differentially expressed after TAp73 was knocked down in the presence of oxaliplatin. A spectrum of miRNAs including miR-519d, miR-545, miR-618 and miR-98 were found to be enhanced in both cell lines, while miR-132, miR-518f and miR-758 were highly expressed specifically in the HCT116p53+/+ cells and correspondingly miR-127 and miR-654 were elevated in the HCT116p53-/- cells. However, these were not observed to be up-regulated after TAp73 was knocked down. In contrast, miR-433 and miR-379 which were minimized in both the cell lines and miR-205 which was specifically down-regulated only in HCT116p53-/- cells were not found to be down-regulated after TAp73 knockdown.
Putative target genes prediction of dysregulated miRNAs
The biochemical function of the differentially expressed miRNAs after oxaliplatin treatment was investigated by predicting the miRNA target genes using two programs: TargetScan Release 6.2 (http://www.targetscan.org) and MicroCosm Targets Version 5. Since the prediction algorithms of these two programs are different, their predicted target genes were also notably different. Many of the largely predicted target genes of most miRNAs that are affected by oxaliplatin treatment were found to be associated with processes involved in the response to genotoxic stress, including the apoptotic process (GO: 0006915), cell death (GO: 0008219), cell cycle (GO: 0007049), cell cycle arrest (GO: 0007050), response DNA damage stimulus (GO: 0006974) and DNA repair (GO: 000628) in gene ontology (GO) (http://www.geneontology.org).
Discussion
The undertaken study aimed to explore the efficacy of oxaliplatin treatment for studying the miRNA expression in colon cancer cells under the influence of activated or inactivated p53 and p73 protein. HCT116p53+/+ with wild-type p53 and HCT116p53-/-, a variant cell line with truncated p53, were utilized as an experimental model to study the effects of the oxaliplatin, a potent chemotherapeutic drug. We focused on the effects of oxaliplatin on the expression of 379 miRNAs after activated/inactivated p53 and p73. After treating both the cells with oxaliplatin dose-dependent reduced cell viability was observed in p53 wild-type cells as compared to p53 negative cells. Both up- and down- regulated expression of several miRNAs were observed in response to oxaliplatin treatment. Interestingly, majority of the miRNAs that were found to be deregulated by oxaliplatin have identified to be associated with the pathogenesis of cancer. After treatment with oxaliplatin, the HCT116p53+/+ cells demonstrated a two-fold increase of about 16 miRNAs whereas in the HCT116p53-/- cells 15 miRNAs were up-regulated. Among the expressed miRNAs, around 11 of the up-regulated miRNAs were found to be common to both the cell lines. In particular, miR-542-3p, miR-511 and miR-98 which have been previously reported [23-25] to be down-regulated and possess tumor suppressive properties in several cancer types which have displayed enhanced expression post-oxaliplatin treatment. Additionally, some miRNAs including miR-519d, miR-142-5p and miR-143 have also been reported to be associated with chemotherapeutic response or chemosensitivity [25]. Report suggests that in HCC cells, miR-519d increase the apoptotic potentials in response to doxorubicin treatment [26]. In another study, miR-142-5p was observed to indicate a prognostic response to the cytotoxic drug gemcitabine in pancreatic cancer patients, where in the manifestation of miR-142-5p was found to be associated with longer survival rate [27]. Previous published literatures have also mentioned the significant role of miR-143 in chemo response and its characteristic function as a putative tumor suppressor in colorectal cancer [28]. Further, miR-143 expression elevates response of chemo-sensitivity to the oxaliplatin drug in colon cancer cells [29].
A, B, C, D Effect of oxaliplatin on the relative expression of several miRNA in colon cancer cell lines HCT116p53+/+ and HCT116p53-/-. miRNAs have the potential to act as molecular markers or even novel targets for treatment, and an increased understanding of the role of miRNAs in chemo response might aid in improving the efficacy of oxaliplatin treatment.
In the present study, some miRNAs were observed to be deregulated as a result of exposure to oxaliplatin treatment in the HCT116p53+/+ or the HCT116p53-/- cells, which is indicative of the characteristic function of p53 in response to chemotherapy. Studies reported that p53 acts as a significant molecular effector of oxaliplatin activity in the cancer cells during the treatment, whereas the p53-mutant cells exhibited high resistance to oxaliplatin treatment when compared to p53 wild-type cells [15]. Recent research emphasized on the role of p53 as a transcription factor which regulates the expression of genes related to cell cycle arrest, apoptosis and DNA repair [30-32], as well as a considerable number of miRNAs dysregulation [16]. It is therefore acceptable to hypothesize that modified expression of certain miRNAs mediates some effects of the p53 response and thereby regulated the oxaliplatin treatment outcome. Considerably, a spectrum of miRNAs such as miR-34c, miR-132, miR-518f, miR-758, and miR-521, were observed to be exclusively up-regulated in the HCT116p53+/+ cells whereas in the HCT116p53-/-cells miR-18b, miR-204, miR-127 and miR-654-3p were up-regulated. Studies have found that p53 directly enhances the expression of miR-34-family and also it is up-regulated when a cell undergoes stress such as DNA-damage in p53 wild-type cells, where it utilizes anti-proliferative effects [33]. Furthermore, in colon cancer cells the expression of miR-34c have been found to display decreased expression [34] along with increased chemo sensitivity to cisplatin in endometrial cancer [35]. miR-132 have been claimed to increase sensitivity to both chemoradiotherapy and radiation in colon cancer cells [36]. In our study, miR-18b were found to be up-regulated in HCT116p53-/- cells, which targets the MDM2-p53 pathway directly and previously proposed to act as an anti-oncogene in melanoma by restoring the tumor suppressive effects of p53 that sensitizes melanoma cells against cisplatin cytotoxicity [37]. Furthermore, miR-204 have the property of targeting the anti-apoptotic protein Bcl-2, thereby increasing the sensitivity to cisplatin in neuroblastoma cells [38]. It is also found to be sensitive to oxaliplatin and 5-fluorouracil in gastric cancer [39]. miR-127 is predominantly found to be down-regulated in cancer cells and also in primary human tumors, and it is suggested to act as a tumor suppressor by inhibiting the proto-oncogene Bcl-6 [40]. Additionally, miR-127 is observed to exhibit reduced expression in doxorubicin-resistant colon cancer cell lines [41].
Selective miRNAs such as miR-570, miR-331-5p, miR-503 and miR-548a displayed reduced expression exclusively in HCT116p53+/+ cells, and miR-205 and miR-153 were found to be down-regulated in HCT116p53-/- cells only. The p53-family was found to predominantly regulate the processing of diverse miRNAs, including miR-503 [42]. In addition, they have been shown to be up-regulated in chemo-resistant ovarian cancer cells [43]. Some other studies have found miR-331-5p to be down-regulated in leukemia and also concomitant with most chemotherapy resistance [44]. miR-153 found to be up-regulated in colorectal cancer cells where it increased the invasiveness and resistance to cisplatin and oxaliplatin [45].
A myriad of miRNAs was noted to be up-regulated in response to oxaliplatin in this study, the consistent inclination is that they are commonly down-regulated in cancer, and therefore act as key factors and play consistent tumor suppressive roles and increased chemo sensitivity. However, the opposite pattern was not found for the miRNAs that were observed to be down-regulated after oxaliplatin treatment. Some of the down-regulated miRNAs in the cancer cells execute the property of tumor suppressors, for example miR-125a-3p [46], miR-433 [47] and miR-379 [48]. miRNAs including miR-367 and miR-215 which have been analyzed in this study were found to exhibit oncogenic properties. miR-367 is up-regulated in lung tumor tissue and found to be associated with an unfavorable prognosis of tumor [49]. In Wilms' tumor blastema, miR-367 perceived to be more highly expressed in tumors that exhibited a poor response to chemotherapy than in tumors with better response [50]. In a study where chemoradiotherapy in rectal cancer patients was completely analyzed, miR-215 was over-expressed in non-responders [51]. Studies have indicated that another imperative miRNA, miRNA-215 whose level of expression is found to be high in cancer tissues are closely associated with poor overall survival of colon cancer patients [52].
However, these seemingly contradictory findings could be explained by the dual behavior of some miRNAs in cancer cells as they seem to act as either oncogenes or tumor suppressors depending on tissue type and specific targets [53]. The effects of a single miRNA likely rely on the expression level of each target mRNA, which varies depending on the cellular context. Consequently, miRNAs often appeared to have the opposite effects towards similar anticancer agents in different cancer types [54].
p53 a significant tumor suppressor gene is well known for its characteristic regulatory function of expression of a wide range of miRNAs all together, however only a little is known regarding p73 which is a homologue of p53 in this context. TAp73 cited as the pro-apoptotic isoform of p73 was silenced using siRNA before the cells were exposed to oxaliplatin in order to monitor whether TAp73 could determine the expression of miRNAs in a similar way as p53. We found that a major proportion of miRNAs that were dysregulated after the drug treatment in the presence of TAp73, were still observed to be dysregulated subsequent to knock-down of TAp73. However, there are some exceptions, implicating that TAp73 could be necessary for the regulation of some miRNAs. Among the expressed miRNAs, we found miR-205, which was decreased in HCT116p53-/- cells after oxaliplatin treatment probably be a target of p73 [55], and we found that the expression level was undetectable in both cell lines with and without oxaliplatin treatment on inhibition of the TAp73 gene.
We have identified that the drug oxaliplatin modulates the miRNA gene expression profile, indicating that miRNAs influence the pharmacodynamic mechanisms of oxaliplatin in colon cancer, especially as many of the deregulated miRNAs are related to cancer and/or chemo response. Both p53 and TAp73 status was also found to modulate the miRNA expression profiles after oxaliplatin chemotherapy. The modulation was partly different from that of p53, indicative of a miRNA regulatory role of p73 in part distinct from p53.
Conclusion
This study shows that miRNAs could mediate the effects of oxaliplatin, and that p53 and TAp73 could also partly regulate the role of miRNAs in chemosensitivity. The results indicated that 1) miRNA profile is associated with effects of oxaliplatin chemotherapy 2) Both p53 and p73 are involved in the regulations of miRNA expression in body.
Acknowledgements
This study was supported by grants from the Swedish Cancer Foundation, Swedish Research Council and the Health Research Council in the South-East of Sweden. The authors thank Jessica Carlsson, Sathya Priya Thandavan and Suhanya Veronica Prasad for her advice and help on the data analysis.
Authors Contributions
JE participated in the design of the study, carried out the experiments and drafted the manuscript. SP contributed to the design of the study and participated in experimental procedures and writing the MS. XFS and HZ conceived of the study, participated in the design and helped in the drafting of the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL. et al. Global cancer statistics, 2012. CA. Cancer J Clin. 2015;65:87-108
2. Giacchetti S, Perpoint B, Zidani R. et al. Phase III multicenter randomized trial of oxaliplatin added to chronomodulated fluorouracil-leucovorin as first-line treatment of metastatic colorectal cancer. J Clin Oncol. 2000;18:136-147
3. Ahmad S. Platinum-DNA interactions and subsequent cellular processes controlling sensitivity to anticancer platinum complexes. Chem Biodivers. 2010;7:543-66
4. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297
5. Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2007;96(Suppl):R40-44
6. Calin GA, Sevignani C, Dumitru CD. et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. PNAS. 2004;101:2999-3004
7. Bandres E, Cubedo E, Agirre X. et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29
8. Zhang B, Pan X, Cobb GP. et al. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1-12
9. Blower PE, Chung JH, Verducci JS. et al. MicroRNAs modulate the chemosensitivity of tumor cells. Mol Cancer Ther. 2008;7:1-9
10. Schee K, Fodstad Ø, Flatmark O. MicroRNAs as Biomarkers in Colorectal Cancer. Am J Pathol. 2010;177:1592-1599
11. Johnstone RW, Ruefli AA, Lowe SW. Apoptosis: a link between cancer genetics and chemotherapy. Cell. 2002;108:153-164
12. Barak Y. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12(2):461-8
13. Arango D, Wilson AJ, Shi Q. et al. Molecular mechanisms of action and prediction of response to oxaliplatin in colorectal cancer cells. Br J Cancer. 2004;91:1931-1946
14. Irwin MS, Kondo K, Marin MC. et al. Chemosensitivity linked to p73 function. Cancer Cell. 2003;3:403-410
15. Toscano F, Fajoui ZE, Gay F. et al. p53 dependent and independent sensitivity to oxaliplatin of colon cancer cells. Biochem Pharmacol. 2007;74:392-406
16. Jost CA, Marin MC, Kaelin WG. p73 is a simian (correction of human) p53- related protein that can induce apoptosis. Nature. 1997;389(6647):191-4
17. Xi Y, Shalgi R, Fodstad O. et al. Differentially regulated micro-RNAs and actively translated messenger RNA transcripts by tumor suppressor p53 in colon cancer. Clin Cancer Res. 2006;12:2014-2024
18. Bunz F, Dutriaux A, Lengauer C. et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497-1501
19. Gentleman RC, Carey VJ, Bates DM. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80
20. Mar JC, Kimura Y, Schroder K. et al. Data-driven normalization strategies for high-throughput quantitative RT-PCR. BMC Bioinformatics. 2009;10:110
21. Jost CA, Marin MC, Kaelin WG Jr. p73 is a simian [correction of human] p53-related protein that can induce apoptosis. Nature. 1997;389:191-194
22. Ishimoto O, Kawahara C, Enjo K. et al. Possible oncogenic potential of DeltaNp73: a newly identified isoform of human p73. Cancer Res. 2002;62:636-641
23. Yoon S, Choi YC, Lee S. et al. Induction of growth arrest by miR-542-3p that targets survivin. FEBS Lett. 2010;584:4048-4052
24. Zhang C, Chi YL, Wang PY. et al. miR-511 and miR-1297 inhibit human lung adenocarcinoma cell proliferation by targeting oncogene TRIB2. PloS one. 2012;7(e):46090
25. Siragam V, Rutnam ZJ, Yang W. et al. MicroRNA miR-98 inhibits tumor angiogenesis and invasion by targeting activin receptor-like kinase-4 and matrix metalloproteinase-11. Oncotarget. 2012;3:1370-1385
26. Fornari F, Milazzo M, Chieco P. et al. In hepatocellular carcinoma miR-519d is up-regulated by p53 and DNA hypomethylation and targets CDKN1A/p21, PTEN, AKT3 and TIMP2. J Pathol. 2012;227:275-285
27. Ohuchida K, Mizumoto K, Kayashima T. et al. MicroRNA expression as a predictive marker for gemcitabine response after surgical resection of pancreatic cancer. Ann Surg Oncol. 2011;18:2381-2387
28. Kitade Y, Akao Y. MicroRNAs and their therapeutic potential for human diseases: microRNAs, miR-143 and -145, function as anti-oncomirs and the application of chemically modified miR-143 as an anti-cancer drug. J Pharmacol Sci. 2010;114:276-280
29. Qian X, Yu J, Yin Y. et al. MicroRNA-143 inhibits tumor growth and angiogenesis and sensitizes chemosensitivity to oxaliplatin in colorectal cancers. Cell Cyc. 2013;12:1385-1394
30. Lowe SW, Schmitt EM, Smith SW. et al. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847-849
31. Brugarolas J, Chandrasekaran C, Gordon JI. et al. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552-557
32. Smith ML, Ford JM, Hollander MC. et al. p53-mediated DNA repair responses to UV radiation: studies of mouse cells lacking p53, p21, and/or gadd45 genes. Mol Cell Biol. 2000;20:3705-3714
33. He L, He X, Lim LP. et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130-1134
34. Toyota M, Suzuki H, Sasaki Y. et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123-4132
35. Jiang L, Meng W, Zeng J. et al. MiR-34c oligonucleotide enhances chemosensitivity of Ishikawa cell to cisplatin by inducing apoptosis. Cell Biol Int. 2013;37:577-583
36. Salendo J, Spitzner M, Kramer F. et al. Identification of a microRNA expression signature for chemoradiosensitivity of colorectal cancer cells, involving miRNAs-320a, -224, -132 and let7g. Radiother Oncol. 2013;108:451-457
37. Dar AA, Majid S, Rittsteuer C. et al. The role of miR-18b in MDM2-p53 pathway signaling and melanoma progression. JNCI. 2013;105:433-442
38. Ryan J, Tivnan A, Fay J. et al. MicroRNA-204 increases sensitivity of neuroblastoma cells to cisplatin and is associated with a favourable clinical outcome. Br J Cancer. 2012;107:967-976
39. Sacconi A, Biagioni F, Canu V. et al. miR-204 targets Bcl-2 expression and enhances responsiveness of gastric cancer. Cell Death Dis. 2012;3:e423
40. Saito Y, Liang G, Egger G. et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435-443
41. Qu J, Zhao L, Zhang P. et al. MicroRNA-195 chemosensitizes colon cancer cells to the chemotherapeutic drug doxorubicin by targeting the first binding site of BCL2L2 mRNA. J Cell Physiol. 2013;230:535-545
42. Boominathan L. The tumor suppressors p53, p63, and p73 are regulators of microRNA processing complex. PloS one. 2010;5:e10615
43. Park YT, Jeong JY, Lee MJ. et al. MicroRNAs overexpressed in ovarian ALDH1-positive cells are associated with chemoresistance. J Ovarian Res. 2013;6:18
44. Feng DD, Zhang H, Zhang P. et al. Down-regulated miR-331-5p and miR-27a are associated with chemotherapy resistance and relapse in leukaemia. J Cell Mol Med. 2011;15:2164-2175
45. Zhang L, Pickard K, Jenei V. et al. miR-153 Supports Colorectal Cancer Progression via Pleiotropic Effects That Enhance Invasion and Chemotherapeutic Resistance. Cancer Res. 2013;73:6435-6447
46. Hashiguchi Y, Nishida N, Mimori K. et al. Down-regulation of miR-125a-3p in human gastric cancer and its clinicopathological significance. Int J Oncol. 2012;40:1477-1482
47. Guo LH, Li H, Wang F. et al. The Tumor Suppressor Roles of miR-433 and miR-127 in Gastric Cancer. Int J Mol Sci. 2013;14:14171-14184
48. Laddha SV, Nayak S, Paul D. et al. Genome-wide analysis reveals downregulation of miR-379/miR-656 cluster in human cancers. Biol Direct. 2013;8:10
49. Campayo M, Navarro A, Vinolas N. et al. Low miR-145 and high miR-367 are associated with unfavourable prognosis in resected nonsmall cell lung cancer. Eur Respir J. 2013;41:1172-1178
50. Watson JA, Bryan K, Williams R. et al. miRNA profiles as a predictor of chemoresponsiveness in Wilms' tumor blastema. PloS one. 2013;8:e53417
51. Svoboda M, Sana J, Fabian P. et al. MicroRNA expression profile associated with response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer patients. Rad Oncol. 2012;7:195
52. Karaayvaz M, Pal T, Song B. et al. Prognostic significance of miR-215 in colon cancer. Clin Colorectal Cancer. 2011;10:340-347
53. Fabbri M, Ivan M, Cimmino A. et al. Regulatory mechanisms of microRNAs involvement in cancer. Expert Opin Biol Ther. 2007;7:1009-1019
54. Blower PE, Verducci JS, Lin S. et al. MicroRNA expression profiles for the NCI-60 cancer cell panel. Mol Cancer Therapeutics. 2007;6:1483-1491
55. Alla V, Kowtharapu BS, Engelmann D. et al. E2F1 confers anticancer drug resistance by targeting ABC transporter family members and Bcl-2 via the p73/DNp73-miR-205 circuitry. Cell Cycle. 2012;11:3067-3078
Author contact
![]() Corresponding author: Hong Zhang, School of Medical Sciences, Örebro University, S-701 82, Örebro, Sweden, E-mail: hong.zhangse
Corresponding author: Hong Zhang, School of Medical Sciences, Örebro University, S-701 82, Örebro, Sweden, E-mail: hong.zhangse

 Global reach, higher impact
Global reach, higher impact