3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(11):1966-1972. doi:10.7150/jca.24550 This issue Cite
Review
Involvement of non-coding RNAs in chemotherapy resistance of ovarian cancer
1. Cancer Institute, Fudan University Shanghai Cancer Center;
2. Department of Oncology, Shanghai Medical College, Fudan University, Shanghai 200032, China;
3. Central Laboratory, The Fifth People's Hospital of Shanghai Fudan University, Shanghai 200240, China
Received 2017-12-27; Accepted 2018-2-25; Published 2018-4-30
Abstract
Ovarian cancer (OC) is the most lethal gynecological malignancy, with a low 5-year survival rate. Most patients with ovarian cancer are diagnosed in late-stages. A rising number of non-coding RNAs (ncRNAs) have been found to act as key regulators of gene expression by applying novel high-thought methods, such as next generation sequencing (NGS). Non-coding RNAs not only play important roles in carcinogenesis, but also affect the clinical treatment strategies. One of the biggest challenge in OC treatment was chemoresistance, which causes poor prognosis and high recurrence rate after applying traditional remedies. Of note, it has been proved that ncRNAs were deeply correlated with chemoresistance in several cancers, which made ncRNAs considered to be potential therapeutic targets in ovarian cancer. Among of all ncRNAs, the studies of miRNAs and lncRNAs in ovarian cancer chemoresistance were much clearer. In this study, we reviewed the most relevant researches in this field, and described the relationships between ncRNAs and chemoresistance in ovarian cancer.
Keywords: ncRNAs, ovarian cancer, chemoresistance, miRNAs, lncRNAs
Introduction
As one of the most malignant cancers, ovarian cancer, especially epithelial ovarian cancer, is the most lethal cancer in gynecological malignancy[1, 2]. The low 5-year survival rate of ovarian cancer, which ranges from 30% to 90%, is the major concern in clinical practice. Over 90% of ovarian cancers are classified as epithelial ovarian cancer. Patients at early stages usually have a high survival rate of nearly 90%, however, a large number of patients with ovarian cancer are diagnosed as late-stage diseases[3, 4]. The methods of diagnosis mainly involve pelvic examination, transvaginal ultrasound, and serum cancer antigen 125 (CA125) measurements, and common therapies for ovarian cancer are surgery combined with neoadjuvant chemotherapy[5]. Over the past decades, although there is a growing understanding of ovarian cancer, the early diagnosis rate and prognosis for the disease were still poor. A large number of studies revealed that non-coding RNAs (ncRNAs) played important roles in regulating various biological processes of ovarian cancer, including chemoresistance[6, 7].
Chemoresistance, which was classified into intrinsic and acquired chemoresistance, is a common phenomenon in chemotherapy. Severe chemoresistance could accelerate cancer progression and induce morality. Intrinsic resistance exists in a part of chemo-naive patients, which causes the ineffectiveness of intended chemotherapy. While acquired resistance is usually emerged when tumors are resistant to a particular drug during the treatment [8].
With the development of next-generation sequencing (NGS), it has been proposed that chemotherapy resistance was closely associated with the dysregulation of genes. Emerging studies had showed ncRNAs played an important role in chemotherapy resistance, which indicated the potential roles of lncRNAs in reducing chemoresistance in the ovarian cancers [8, 9].
NcRNAs are RNA molecules that are not translated into protein, including highly abundant and functionally important RNAs such as transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as microRNAs, lncRNAs, circRNAs and tRFs[10]. They have been proved to regulate many biological processes, such as cancer prognosis, response to chemotherapy and radiotherapy sensitivity. Dysregulation of ncRNAs in chemotherapy-sensitive cells suggests that chemotherapy resistance could be regulated by regulating ncRNA expression[11-14]. Through participating in several signaling pathways, such as TGF-β/SMAD[15] and p53 signaling pathway[16], ncRNAs can be developed to serve as potential diagnostic and prognostic biomarkers in ovarian cancer.
NcRNAs involved in chemoresistance of OC
Increased intracellular miRNA expression could affect chemoresistance through epigenetic changes. Increasing evidences demonstrated that various miRNAs and lncRNAs are involved in OC chemoresistance[17, 18].
Chemoresistance mechanisms in OC
Optimal treatment for advanced ovarian cancer includes surgical cytoreduction and platinum-based chemotherapy. A recent study demonstrated that patients with stages III and IV ovarian cancer who received chemotherapy had non-inferior survival rate and reduced treatment-related mortality compared with those who received primary cytoreductive surgery[19]..
Nearly 20% of ovarian cancer patients are resistant to the standard platinum-based chemotherapy, which made chemoresistance to be one of the most critical problems in ovarian cancer therapy. Patients treated with chemotherapy not only need to face with recrudesce of the disease but also have to take the risk of drug resistance. What's more, there is usually no effective curative treatment for acquired resistance yet [20, 21].
Recent studies demonstrate that there are several leading causes to induce chemoresistance. The first and maybe the most important is the pharmacokinetic profile of the drugs. Efflux happens in the transportation progress of many drugs before they could reach the therapeutic targets, and then causes the reduction of drug absorptions and curative effects. The second is the drug inactivation. Although some drugs could reach the therapeutic targets, they couldn't be activated and take effect, which will cause resistance. This mechanism is found to be closely connected with drug-induced DNA damage repair. The last is the reactivation of the targeted pathway. Drugs successfully reach the therapeutic targets and take effect, but they fail to cause subsequent reactions and kill cancer cells. This process is thought as the results of cross-talk between microenvironment, apoptosis inhibition, autophagy induction and alterations in cell cycle checkpoints[22].
Impact of ncRNAs on chemoresistance in OC
Compared with other ncRNAs, the functions and regulation mechanisms of miRNAs were well studied. A transcriptomic study performed in different ovarian cancer cells found that let-7e, miR-30c, miR-125b, miR-130a and miR-335 had abnormal expressions in all the drug resistant cells, including cisplatin, paclitaxel- and cyclosporin A [23].
In a series of studies, miRNAs have been verified to be correlated with chemotherapy resistance in ovarian cancer. Let-7i was the first miRNAs which was thought to be closely associated with chemotherapy-resistant EOC. The decreased levels of let-7i was observed in chemotherapy-resistant EOC patients with shorter survival rate. In a related study, resistance to cisplatin was increased by inhibition of let-7i expression in ovarian cancer [24].
MiR-106a and miR-591 were proved to play roles in paclitaxel resistance in both ovarian cancer cells and tumor samples. Overexpression of anti-miR-106a or pre-miR-591 could recover the sensitivity of paclitaxel-resistant cells to paclitaxel by activating apoptosis and inhibiting cell migration and proliferation [25]. Downregulation of miR-106a increased the protein levels of RB1, p21, caspase-7 and BCL10, meanwhile upregulation of miR-591 suppressed ZEB1 expression.
A systematic study of miRNA demonstrated that a series of miRNAs were associated with paclitaxel resistance in ovarian cancer cells, including miR-663, miR-622, miR-497, miR-187, miR-195 and miR-107. The regulation processes of these miRNAs were mainly related to p53 networks[26].
MiR-21, which plays crucial roles in cancer progression, has also been proved to be associated with OC resistance. Yu et al reported that miR-21 could enhance the chemoresistance to cisplatin through suppressing PTEN expression in epithelial ovarian cancer[27]. Besides, compared with ovarian cancer cells, miR-21 isomiRNAs had higher levels in exosomes and tissue lysates isolated from ovarian cancer-associated adipocytes (CAAs) and fibroblasts (CAFs). They could transferred to ovarian cancer cells from CAAs and CAFs, and therefore enhance OC paclitaxel-resistance by binding to APAF1[28]. This implies that chemoresistance occurred in cancer cells induced by miRNAs may not be the results of in situ abnormal expression of miRNAs but rather the transference of stromal-derived miRNAs. Additionally, miR-21 was found to be related with chemoresistance in other gynecologic cancers, including breast cancer[29] and cervical cancer[30]. The complex mechanisms of miR-21 mediated chemoresistance deserve deeper consideration in future research.
A system biology revealed that, in cisplatin-resistant OC, four candidate miRNAs may have great contributions, including miR-24-3p, miR-192-5p, miR-139-5p and miR-155-5p. The functions of these miRNAs were closely related with apoptosis, adhesion and cell cycle[31].
Some miRNAs also made important functions in paclitaxel resistance in OC. Upregulation of miR-136 could resensitize paclitaxel-resistant by targeting Notch3 in OC cells [32]. Upregulation of miR-1307 could inhibit ING5 levels and promote paclitaxel chemoresistance in OC [33]. MiR-34c-5p could suppress docetaxel and carboplatin resistance in OC via targeting AREG and downregulating the AREG-EGFR-ERK pathway[34].
Six miRNAs, including miR-23b, miR-27a, miR-27b, miR-346, miR-424 and miR-503, were upregulated in ALDH1-positive ovarian cells, and they all had high expression in chemoresistant ovarian cancer cells and tumor samples compared with chemosensitive group. High ALDH1 expression was associated with chemoresistance both in vitro and in vivo [35].
Besides, a series of miRNAs were also reported to have associations with cisplatin and paclitaxel resistance in OC (Table 1). MiR-200 family could affect sensitivities to cisplatin and paclitaxel by targeting KEAP1[36], p38α[37], ZEB1[38] and TUBB3[39]. MiR-199/214 cluster could induce or suppress cisplatin and paclitaxel resistance through inhibiting several target genes, including IKKβ[40], HIF-1A[40], HIF-2A[40], CD44[41], JAG1[42], PTEN[43] and CCL5[43]. MiR-31 could function as a paclitaxel-resistance suppressor by directly targeting MET [44].
Among of all ncRNAs, the mechanisms of miRNAs in chemoresistance were the most clear. Through directly targeting genes or interacting with other ncRNAs, miRNAs could affect paclitaxel, cisplatin and carboplatin resistance in ovarian cancer. By this way, these miRNAs were involved in numerous signaling pathways, and formed large-scale regulatory networks[45]. Except mentioned miRNAs, some others also played roles in OC chemoresistance, such as miR-146a[46], miR-9[47] and miR-409-3p[48].
MicroRNAs and chemotherapy resistance in ovarian cancer
| miRNAs | Target genes | Resistance | Reference | |
|---|---|---|---|---|
| let-7i | N/A | Cisplatin resistance | [24] | |
| miR-106a | RB1, p21, BCL10, Caspase-7 | Paclitaxel resistance | [25] | |
| miR-591 | ZEB1 | Paclitaxel resistance | [25] | |
| miR-663 | p53 networks | Paclitaxel resistance | [26] | |
| miR-622 | p53 networks | Paclitaxel resistance | [26] | |
| miR-497 | p53 networks | Paclitaxel resistance | [26] | |
| miR-187 | p53 networks | Paclitaxel resistance | [26] | |
| miR-195 | p53 networks | Paclitaxel resistance | [26] | |
| miR-107 | p53 networks | Paclitaxel resistance | [26] | |
| miR-24-3p | N/A | Cisplatin resistance | [31] | |
| miR-192-5p | N/A | Cisplatin resistance | [31] | |
| miR-139-5p | N/A | Cisplatin resistance | [31] | |
| miR-155-5p | N/A | Cisplatin resistance | [31] | |
| miR-21 | PTEN | Cisplatin resistance | [27] | |
| APAF1 | Paclitaxel resistance | [28] | ||
| miR-136 | Notch3 | Paclitaxel resistance | [32] | |
| miR-1307 | ING5 | Paclitaxel resistance | [33] | |
| miR-34c-5p | AREG | Docetaxel/Carboplatin resistance | [34] | |
| miR-23b | ALDH1 | Pactitaxel resistance | [5] | |
| miR-27a | ALDH1 | Pactitaxel resistance | [35] | |
| miR-27b | ALDH1 | Pactitaxel resistance | [35] | |
| miR-346 | ALDH1 | Pactitaxel resistance | [35] | |
| miR-424 | ALDH1 | Pactitaxel resistance | [35] | |
| miR-503 | ALDH1 | Pactitaxel resistance | [35] | |
| miR-200 family | miR-141 | KEAP1 | Cisplatin resistance | [36] |
| miR-141,miR-200a | p38α | Pactitaxel resistance | [37] | |
| miR-200c | ZEB1 | Pactitaxel resistance | [38] | |
| miR-200c | TUBB3 | Pactitaxel resistance | [39] | |
| miR-199/214 cluster | miR-199a-5p | IKKβ, HIF-1A, HIF-2A | Paclitaxel resistance | [40] |
| miR-199a-3p | CD44 | Cisplatin/Pactitaxel resistance | [41] | |
| miR-199b-5p | JAG1 | Cisplatin resistance | [42] | |
| miR-214 | PTEN, CCL5 | Cisplatin resistance | [43] | |
| miR-31 | MET | Paclitaxel resistance | [44] |
In present studies, a series of miRNAs were considered as potential diagnostic and prognostic biomarkers in chemoresistance. Due to the abundance of miRNAs, it was believed that the accuracy of diagnosis in OC chemoresistance could be elevated by integrating different kinds of miRNAs. A series of miRNAs, including Miravirsen, RG-101, MRX34, MesomiR-1, MRG-201, MRG-106 and RG-125/ AZD4076, have been or are being tested for therapy in clinical trials in different disease areas. Although most of them are not designed for cancer chemoresistance, it is hopeful that some miRNAs can be as drug targets in chemoresistance treatment in the future.
An increasing number of lncRNAs were proved to play key roles in OC chemoresistance (Table 2). HOX antisense intergenic RNA (HOTAIR), a kind of classical lncRNA, was found to play important roles in relating DNA damage with chemoresistance in ovarian cancer[49]. In addition, HOTAIR could also activate NF-κB signaling pathway. In a relevant study, upregulation of HOTAIR could induce platinum resistance in ovarian cancer, and the expression level of HOTAIR was significantly increased in recurrent platinum-resistant ovarian tumors. This suggested that NF-κB-HOTAIR axis could influence DNA damage and cause chemoresistance through driving a positive-feedback loop in a series of cancers[50]. In other study, through activating wnt/β-catenin signaling pathway, the overexpression of HOTAIR could promote cell cycle progression and induce cellular cisplatin resistance[51]. However, in other gynecologic cancers, the relationship between HOTAIR and chemoresistance is still unclear.
BC200 was also reported to be related with ovarian cancer. It was observed that carboplatin could upregulate the expression of BC200 in cell lines, and the drug sensitivity of the cells was suppressed when inhibiting the level of BC200. So, BC200 appears to serve as a regulator of carboplatin-induced ovarian cancer cell death[52].
NEAT1, another important lncRNA, was proved to affect paclitaxel resistance in ovarian cancer. A related study demonstrated that NEAT1 was overexpressed in paclitaxel-resistant ovarian cancer tissues. NEAT1 could upregulate ZEB1 by sponging miR-194, and knockdown of NEAT1 could enhance the sensitivity of ovarian cancer cells to paclitaxel [53].
UCA1, related to SPRK1, was found to affect cisplatin resistance in ovarian cancer. The expression of UCA1 could enhance cisplatin resistance, and knocking-down SRPK1 could partly rescue the effect of UCA1 on cisplatin resistance[54].
In Midkine (MK)-related ovarian cancer, lncRNA ANRIL may function as a risk factor. The knock-down of ANRIL suppressed MK-induced cisplatin resistance and caused apoptosis via activating caspase-3/Bcl-2[55].
At present, the studies of lncRNAs and OC chemoresistance are still at early stage. One of the most common regulatory mechanisms of lncRNAs is to regulate the chemoresistance in ovarian and other cancers by sponging miRNAs [56, 57]. However, only a small quantity of the numerous lncRNAs have been found to be associated with chemoresistance. The related mechanisms only include DNA damage and cell apoptosis, and limited signaling pathways are proved to affect OC chemoresistance, including NF-κB, wnt/β-catenin and cell apoptosis signaling pathway. Further studies are needed to explore more potential lncRNAs with regulatory roles in chemoresistance.
LncRNAs and chemotherapy resistance in ovarian cancer
| LncRNAs | Target miRNAs | Target genes | Resistance | Reference |
|---|---|---|---|---|
| HOTAIR | IκBα | Platinum resistance | [50] | |
| wnt/β-catenin pathway | Cisplatin resistance | [51] | ||
| BC200 | Carboplatin resistance | [52] | ||
| NEAT1 | miR-194 | ZEB1 | Paclitaxel resistance | [53] |
| UCA1 | SRPK1 | Cisplatin resistance | [54] | |
| ANRIL | caspase-3/Bcl-2 | Cisplatin resistance | [55] |
Nowadays, circular RNAs (circRNAs) and tRNA-derived RNA fragments (tRFs) have been developed into new research targets in chemoresistance [58-60]. For now, there hasn't been any reports that proposed any direct relationships between chemotherapy resistance with circRNAs or tRFs. However, Wu et al reported that circRNAs had close relationships with radioresistance in esophageal cancer [61]. tRFs were also proved to regulate several chemoresistance-related genes, including AURKA[62, 63] and YBX1[64, 65]. In addition, it has been reported that cirRNAs and tRFs may play a key role in chemotherapy resistance through altering stress conditions in cancer microenvironment. Above all, circRNAs and tRFs were believed to be the research hotspot in the future.
Chemoresistance and precision medicine
As a result of the development of modern medical technology, pathogenesis of many critical diseases are being uncovered. With the advancement of NGS and HGP, the therapeutic strategies for cancers have gradually turned towards precision medical. More and more studies have demonstrated the critical roles of RNAs in tumor treatment.
Among of all ncRNAs, miRNAs are most widely studied and have the potential to serve as diagnostic biomarkers and the targets for therapeutic drugs. For example, the most famous miRNAs, miR-34, plays important roles in breast cancer and ovarian cancer by targeting EGFR, Bcl-2, CCND1 and some other important genes [66-68]. Compared with other ncRNAs, the functions of miRNAs are much clearer. They usually functioned through participating in several signaling pathways. The characteristic of multifunction make it possible for miRNAs to have a pivotal role in precision medical.
As a kind of novel ncRNAs, researches on tRFs are still at an early stage. Since the similarity with miRNAs in structure, tRFs were considered to be a kind of miRNAs. Besides, generation of tRFs is closely related with stress conditions, which implies that tRFs are more sensitive to cancer microenvironment, and could be developed to a diagnostic biomarker.
A large number of studies that proposed circRNAs can function as ceRNA make it to be hotspots in cancer research[69, 70]. Because of the closed loop structure, circRNAs could hardly be cleaved by exonuclease. Because of this, circRNAs are stable and have long half-lives in cells and human blood. Based on this characteristic, circRNAs are considered as the most suitable biomarker for early diagnosis.
Based on the unique features, lncRNAs can induce gene silencing by combing with other ncRNAs and mRNAs. The biological functions of lncRNAs is complex. In addition to the roles mentioned above, they can even encode proteins. What's more, lncRNAs can function as the bridges for the crosstalk between other ncRNAs.
With the development of precision medical, the relationships between ncRNAs and diseases will be the future interests in many research fields, including cancers and chemotherapy resistance. Researches on all kinds of ncRNAs have made remarkable progress, but it is believed that there still exists other types of ncRNAs that are uncovered, and they can form an intact RNA world (Figure 1).
Future Perspectives
ncRNAs have a variety of roles in cells, including affecting the structure of the whole chromosomes, regulating gene expressions and mediating the translation of genetic codons into protein sequences. ncRNAs can be classified into several types and distinguished from each other according to their different functions, including unique regulatory mechanisms, alternative forms of biogenesis and functional structured RNA domains. Current deep RNA-sequencing and advanced epigenomic technologies could help to identify new ncRNAs and analyze their characteristics. In the future, more features and functions of ncRNAs will be uncovered gradually with the help of future techniques. It can be expected that ncRNA-based therapeutics will play an important role in future clinical practice[71].
Based on present studies, miRNAs are the most studied ncRNAs in ovarian cancer chemoresistance. Several miRNAs have been identified to be remarkably up/down-regulated in chemoresistant ovarian cancer, such as let-7i, miR-27a, miR-23a, miR-449b, miR-21 and so on[72]. These miRNAs were proved to be associated with poor prognosis and short survival in chemoresistant ovarian cancer patients. Of note, the research of miR-21 reveals that, in cancer chemoresistance, we should focus on the diversity of regulatory mechanism and assess the biological function of miRNAs in many aspects, including in situ and transfer.
The relationships between ncRNAs and chemotherapy resistance in ovarian cancer
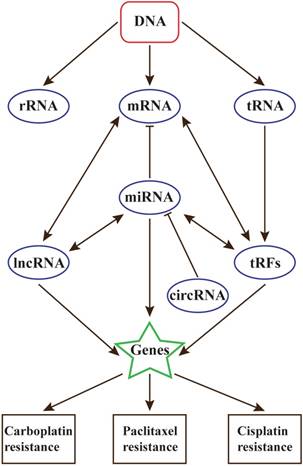
Besides, lncRNA also play key roles in chemoresistance. They are closely linked with DNA damage and repair. However, the present studies of lncRNAs in OC chemoresistance are much limited, and HOTAIR may play the most important role. In near future, the relationship between lncRNAs and chemoresistance will be the highlight of ovarian cancer research.
Contrast to miRNAs and lncRNAs, whether circRNAs and tRFs participate in chemoresistance are still unclear. In view of the relationships between these ncRNAs, circRNAs and tRFs are also believed to contribute to chemoresistance in ovarian cancer[18].
Although breakthroughs have been made on regulatory mechanisms of sncRNAs, there are still a lot of unknown aspects need to be explored. For example, tRFs were affirmed to be a new type of ncRNAs cleaved from tRNAs, rather than a kind of miRNAs as previously considered. So, it is still necessary for us to focus on ncRNAs, not only to correct any possible mistakes, but also to contribute to the development of physianthropy.
Acknowledgements
The authors thank Dr. Zhongliang Ma (Shanghai University, Shanghai, China) for giving advices in the course of this work.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. et al. Cancer statistics in China, 2015. CA: a cancer journal for clinicians. 2016
2. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA: a cancer journal for clinicians. 2017;67:7-30
3. Di Leva G, Croce CM. miRNA profiling of cancer. Current opinion in genetics & development. 2013;23:3-11
4. Prat J, Oncology FCoG. Staging classification for cancer of the ovary, fallopian tube, and peritoneum. International journal of gynaecology and obstetrics: the official organ of the International Federation of Gynaecology and Obstetrics. 2014;124:1-5
5. van Haaften-Day C, Shen Y, Xu F, Yu Y, Berchuck A, Havrilesky LJ. et al. OVX1, macrophage-colony stimulating factor, and CA-125-II as tumor markers for epithelial ovarian carcinoma: a critical appraisal. Cancer. 2001;92:2837-44
6. Redman C, Duffy S, Bromham N, Francis K, Guideline Development G. Recognition and initial management of ovarian cancer: summary of NICE guidance. Bmj. 2011;342:d2073
7. Karlan BY, Alvarez RD, Strauss JF 3rd. Evolving Paradigms in Research and Care in Ovarian Cancers. Obstetrics and gynecology. 2016;128:771-4
8. Frederick PJ, Green HN, Huang JS, Egger ME, Frieboes HB, Grizzle WE. et al. Chemoresistance in ovarian cancer linked to expression of microRNAs. Biotechnic & histochemistry: official publication of the Biological Stain Commission. 2013;88:403-9
9. Xing Z, Li D, Yang L, Xi Y, Su X. MicroRNAs and anticancer drugs. Acta biochimica et biophysica Sinica. 2014;46:233-9
10. Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S. et al. Hidden layers of human small RNAs. BMC genomics. 2008;9:157
11. Xiao J, Lv Y, Jin F, Liu Y, Ma Y, Xiong Y. et al. LncRNA HANR Promotes Tumorigenesis and Increase of Chemoresistance in Hepatocellular Carcinoma. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2017;43:1926-38
12. Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu XY. et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/beta-catenin signaling. Molecular cancer. 2017;16:9
13. Danquah M, Singh S, Behrman SW, Mahato RI. Role of miRNA and cancer stem cells in chemoresistance and pancreatic cancer treatment. Expert opinion on drug delivery. 2012;9:1443-7
14. Buhagiar A, Ayers D. Chemoresistance, cancer stem cells, and miRNA influences: the case for neuroblastoma. Analytical cellular pathology. 2015;2015:150634
15. Parikh A, Lee C, Joseph P, Marchini S, Baccarini A, Kolev V. et al. microRNA-181a has a critical role in ovarian cancer progression through the regulation of the epithelial-mesenchymal transition. Nature communications. 2014;5:2977
16. Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N. et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7004-9
17. Chen QN, Wei CC, Wang ZX, Sun M. Long non-coding RNAs in anti-cancer drug resistance. Oncotarget. 2017;8:1925-36
18. Ayers D, Vandesompele J. Influence of microRNAs and Long Non-Coding RNAs in Cancer Chemoresistance. Genes. 2017:8
19. Vergote I, Trope CG, Amant F, Kristensen GB, Ehlen T, Johnson N. et al. Neoadjuvant chemotherapy or primary surgery in stage IIIC or IV ovarian cancer. The New England journal of medicine. 2010;363:943-53
20. Prahm KP, Novotny GW, Hogdall C, Hogdall E. Current status on microRNAs as biomarkers for ovarian cancer. APMIS: acta pathologica, microbiologica, et immunologica Scandinavica. 2016;124:337-55
21. Weinberg LE, Rodriguez G, Hurteau JA. The role of neoadjuvant chemotherapy in treating advanced epithelial ovarian cancer. Journal of surgical oncology. 2010;101:334-43
22. Fanale D, Castiglia M, Bazan V, Russo A. Involvement of Non-coding RNAs in Chemo- and Radioresistance of Colorectal Cancer. Advances in experimental medicine and biology. 2016;937:207-28
23. Sorrentino A, Liu CG, Addario A, Peschle C, Scambia G, Ferlini C. Role of microRNAs in drug-resistant ovarian cancer cells. Gynecologic oncology. 2008;111:478-86
24. Yang N, Kaur S, Volinia S, Greshock J, Lassus H, Hasegawa K. et al. MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer research. 2008;68:10307-14
25. Huh JH, Kim TH, Kim K, Song JA, Jung YJ, Jeong JY. et al. Dysregulation of miR-106a and miR-591 confers paclitaxel resistance to ovarian cancer. British journal of cancer. 2013;109:452-61
26. Kim YW, Kim EY, Jeon D, Liu JL, Kim HS, Choi JW. et al. Differential microRNA expression signatures and cell type-specific association with Taxol resistance in ovarian cancer cells. Drug design, development and therapy. 2014;8:293-314
27. Yu X, Chen Y, Tian R, Li J, Li H, Lv T. et al. miRNA-21 enhances chemoresistance to cisplatin in epithelial ovarian cancer by negatively regulating PTEN. Oncology letters. 2017;14:1807-10
28. Au Yeung CL, Co NN, Tsuruga T, Yeung TL, Kwan SY, Leung CS. et al. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nature communications. 2016;7:11150
29. Dai X, Fang M, Li S, Yan Y, Zhong Y, Du B. miR-21 is involved in transforming growth factor beta1-induced chemoresistance and invasion by targeting PTEN in breast cancer. Oncology letters. 2017;14:6929-36
30. Feng Y, Zou W, Hu C, Li G, Zhou S, He Y. et al. Modulation of CASC2/miR-21/PTEN pathway sensitizes cervical cancer to cisplatin. Archives of biochemistry and biophysics. 2017;623-624:20-30
31. Liu W, Wang S, Zhou S, Yang F, Jiang W, Zhang Q. et al. A systems biology approach to identify microRNAs contributing to cisplatin resistance in human ovarian cancer cells. Molecular bioSystems. 2017;13:2268-76
32. Jeong JY, Kang H, Kim TH, Kim G, Heo JH, Kwon AY. et al. MicroRNA-136 inhibits cancer stem cell activity and enhances the anti-tumor effect of paclitaxel against chemoresistant ovarian cancer cells by targeting Notch3. Cancer letters. 2017;386:168-78
33. Chen WT, Yang YJ, Zhang ZD, An Q, Li N, Liu W. et al. MiR-1307 promotes ovarian cancer cell chemoresistance by targeting the ING5 expression. Journal of ovarian research. 2017;10:1
34. Tung SL, Huang WC, Hsu FC, Yang ZP, Jang TH, Chang JW. et al. miRNA-34c-5p inhibits amphiregulin-induced ovarian cancer stemness and drug resistance via downregulation of the AREG-EGFR-ERK pathway. Oncogenesis. 2017;6:e326
35. Park YT, Jeong JY, Lee MJ, Kim KI, Kim TH, Kwon YD. et al. MicroRNAs overexpressed in ovarian ALDH1-positive cells are associated with chemoresistance. Journal of ovarian research. 2013;6:18
36. van Jaarsveld MT, Helleman J, Boersma AW, van Kuijk PF, van Ijcken WF, Despierre E. et al. miR-141 regulates KEAP1 and modulates cisplatin sensitivity in ovarian cancer cells. Oncogene. 2013;32:4284-93
37. Mateescu B, Batista L, Cardon M, Gruosso T, de Feraudy Y, Mariani O. et al. miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nature medicine. 2011;17:1627-35
38. Cochrane DR, Spoelstra NS, Howe EN, Nordeen SK, Richer JK. MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Molecular cancer therapeutics. 2009;8:1055-66
39. Cittelly DM, Dimitrova I, Howe EN, Cochrane DR, Jean A, Spoelstra NS. et al. Restoration of miR-200c to ovarian cancer reduces tumor burden and increases sensitivity to paclitaxel. Molecular cancer therapeutics. 2012;11:2556-65
40. Chen R, Alvero AB, Silasi DA, Kelly MG, Fest S, Visintin I. et al. Regulation of IKKbeta by miR-199a affects NF-kappaB activity in ovarian cancer cells. Oncogene. 2008;27:4712-23
41. Cheng W, Liu T, Wan X, Gao Y, Wang H. MicroRNA-199a targets CD44 to suppress the tumorigenicity and multidrug resistance of ovarian cancer-initiating cells. The FEBS journal. 2012;279:2047-59
42. Liu MX, Siu MK, Liu SS, Yam JW, Ngan HY, Chan DW. Epigenetic silencing of microRNA-199b-5p is associated with acquired chemoresistance via activation of JAG1-Notch1 signaling in ovarian cancer. Oncotarget. 2014;5:944-58
43. Yang H, Kong W, He L, Zhao JJ, O'Donnell JD, Wang J. et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer research. 2008;68:425-33
44. Mitamura T, Watari H, Wang L, Kanno H, Hassan MK, Miyazaki M. et al. Downregulation of miRNA-31 induces taxane resistance in ovarian cancer cells through increase of receptor tyrosine kinase MET. Oncogenesis. 2013;2:e40
45. Ragusa M, Barbagallo C, Brex D, Caponnetto A, Cirnigliaro M, Battaglia R. et al. Molecular Crosstalking among Noncoding RNAs: A New Network Layer of Genome Regulation in Cancer. International journal of genomics. 2017;2017:4723193
46. Wilczynski M, Zytko E, Szymanska B, Dzieniecka M, Nowak M, Danielska J. et al. Expression of miR-146a in patients with ovarian cancer and its clinical significance. Oncology letters. 2017;14:3207-14
47. Li X, Pan Q, Wan X, Mao Y, Lu W, Xie X. et al. Methylation-associated Has-miR-9 deregulation in paclitaxel- resistant epithelial ovarian carcinoma. BMC cancer. 2015;15:509
48. Cheng Y, Ban R, Liu W, Wang H, Li S, Yue Z. et al. MiRNA-409-3p enhances cisplatin-sensitivity of ovarian cancer cells by blocking the autophagy mediated by Fip200. Oncology research. 2018
49. Wang Y, Wang H, Song T, Zou Y, Jiang J, Fang L. et al. HOTAIR is a potential target for the treatment of cisplatinresistant ovarian cancer. Molecular medicine reports. 2015;12:2211-6
50. Ozes AR, Miller DF, Ozes ON, Fang F, Liu Y, Matei D. et al. NF-kappaB-HOTAIR axis links DNA damage response, chemoresistance and cellular senescence in ovarian cancer. Oncogene. 2016;35:5350-61
51. Li J, Yang S, Su N, Wang Y, Yu J, Qiu H. et al. Overexpression of long non-coding RNA HOTAIR leads to chemoresistance by activating the Wnt/beta-catenin pathway in human ovarian cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37:2057-65
52. Wu DI, Wang T, Ren C, Liu L, Kong D, Jin X. et al. Downregulation of BC200 in ovarian cancer contributes to cancer cell proliferation and chemoresistance to carboplatin. Oncology letters. 2016;11:1189-94
53. An J, Lv W, Zhang Y. LncRNA NEAT1 contributes to paclitaxel resistance of ovarian cancer cells by regulating ZEB1 expression via miR-194. OncoTargets and therapy. 2017;10:5377-90
54. Wang F, Zhou J, Xie X, Hu J, Chen L, Hu Q. et al. Involvement of SRPK1 in cisplatin resistance related to long non-coding RNA UCA1 in human ovarian cancer cells. Neoplasma. 2015;62:432-8
55. Zhang D, Ding L, Li Y, Ren J, Shi G, Wang Y. et al. Midkine derived from cancer-associated fibroblasts promotes cisplatin-resistance via up-regulation of the expression of lncRNA ANRIL in tumour cells. Scientific reports. 2017;7:16231
56. Du P, Zhao H, Peng R, Liu Q, Yuan J, Peng G. et al. LncRNA-XIST interacts with miR-29c to modulate the chemoresistance of glioma cell to TMZ through DNA mismatch repair pathway. Bioscience reports. 2017:37
57. Shen CJ, Cheng YM, Wang CL. LncRNA PVT1 epigenetically silences miR-195 and modulates EMT and chemoresistance in cervical cancer cells. Journal of drug targeting. 2017;25:637-44
58. Qu S, Yang X, Li X, Wang J, Gao Y, Shang R. et al. Circular RNA: A new star of noncoding RNAs. Cancer letters. 2015;365:141-8
59. Lasda E, Parker R. Circular RNAs: diversity of form and function. Rna. 2014;20:1829-42
60. Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes & development. 2009;23:2639-49
61. Su H, Lin F, Deng X, Shen L, Fang Y, Fei Z. et al. Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. Journal of translational medicine. 2016;14:225
62. Shao Y, Sun Q, Liu X, Wang P, Wu R, Ma Z. tRF-Leu-CAG promotes cell proliferation and cell cycle in non-small cell lung cancer. Chemical biology & drug design. 2017;90:730-8
63. Zhang K, Chen J, Chen D, Huang J, Feng B, Han S. et al. Aurora-A promotes chemoresistance in hepatocelluar carcinoma by targeting NF-kappaB/microRNA-21/PTEN signaling pathway. Oncotarget. 2014;5:12916-35
64. Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell. 2015;161:790-802
65. Guo Y, Pang Y, Gao X, Zhao M, Zhang X, Zhang H. et al. MicroRNA-137 chemosensitizes colon cancer cells to the chemotherapeutic drug oxaliplatin (OXA) by targeting YBX1. Cancer biomarkers: section A of Disease markers. 2017;18:1-9
66. Li N, Fu H, Tie Y, Hu Z, Kong W, Wu Y. et al. miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer letters. 2009;275:44-53
67. Li XJ, Ji MH, Zhong SL, Zha QB, Xu JJ, Zhao JH. et al. MicroRNA-34a modulates chemosensitivity of breast cancer cells to adriamycin by targeting Notch1. Archives of medical research. 2012;43:514-21
68. Zhou JY, Chen X, Zhao J, Bao Z, Chen X, Zhang P. et al. MicroRNA-34a overcomes HGF-mediated gefitinib resistance in EGFR mutant lung cancer cells partly by targeting MET. Cancer letters. 2014;351:265-71
69. Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A. et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017:357
70. Chen J, Cui L, Yuan J, Zhang Y, Sang H. Circular RNA WDR77 target FGF-2 to regulate vascular smooth muscle cells proliferation and migration by sponging miR-124. Biochemical and biophysical research communications. 2017;494:126-32
71. Enfield KS, Martinez VD, Marshall EA, Stewart GL, Kung SH, Enterina JR. et al. Deregulation of small non-coding RNAs at the DLK1-DIO3 imprinted locus predicts lung cancer patient outcome. Oncotarget. 2016
72. Mahdian-Shakib A, Dorostkar R, Tat M, Hashemzadeh MS, Saidi N. Differential role of microRNAs in prognosis, diagnosis, and therapy of ovarian cancer. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2016;84:592-600
Author contact
![]() Corresponding author: E-mail: yanggongedu.cn
Corresponding author: E-mail: yanggongedu.cn

 Global reach, higher impact
Global reach, higher impact