3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(2):375-388. doi:10.7150/jca.21044 This issue Cite
Review
miR-149 in Human Cancer: A Systemic Review
1. The First Clinical School of Nanjing Medical University, Nanjing 210029, P.R. China;
2. Center of Clinical Laboratory, Nanjing Medical University Affiliated Cancer Hospital Cancer Institute of Jiangsu Province, Baiziting 42, Nanjing 210009, P.R. China;
3. Department of General Surgery, the First Affiliated Hospital with Nanjing Medical University, Nanjing 210029, P.R. China.
* Equal contributors
Received 2017-5-16; Accepted 2017-11-21; Published 2018-1-1
Abstract
MicroRNAs (miRNAs) are small noncoding RNAs that regulate post-transcriptional gene expression via binding to the 3'-untranslated region (3'-UTR) of targeted mRNAs. They are reported to play important roles in tumorigenesis and progression of various cancers. Among them, miR-149 was confirmed to be aberrantly regulated in various tumors. In this review, we provide a complex overview of miR-149, particularly summarize the critical roles of it in cancers and expect to lay the foundation for future works on this important microRNA.
Keywords: miRNA, miR-149, cancer.
Introduction
MicroRNAs (miRNAs), a class of highly conserved noncoding RNA (ncRNA) molecules of approximately 22-nucleotide long, are post-transcriptional regulators that bind specifically to the 3′-untranslated region (3′-UTR) of targeted mRNAs, resulting in translational inhibition.1 In the last decade, miRNAs have emerged in many aspects of tumorigenesis, tumor development, metastasis, and drug resistance.2 Among them, miR-149, which can serve as an oncogene (oncomiR) and tumor suppressor, was found to be significantly dysregulated in various cancers, such as head and neck, lung, gastric, breast, prostate cancers and renal cell carcinoma (RCC) (Table 1, 2). In this review, to understand the overview of miR-149 better, we focused on enumerating and summarizing the specific functions and mechanisms of miR-149 in a wide range of cancers.
Structure and chromosomal localization of miR-149
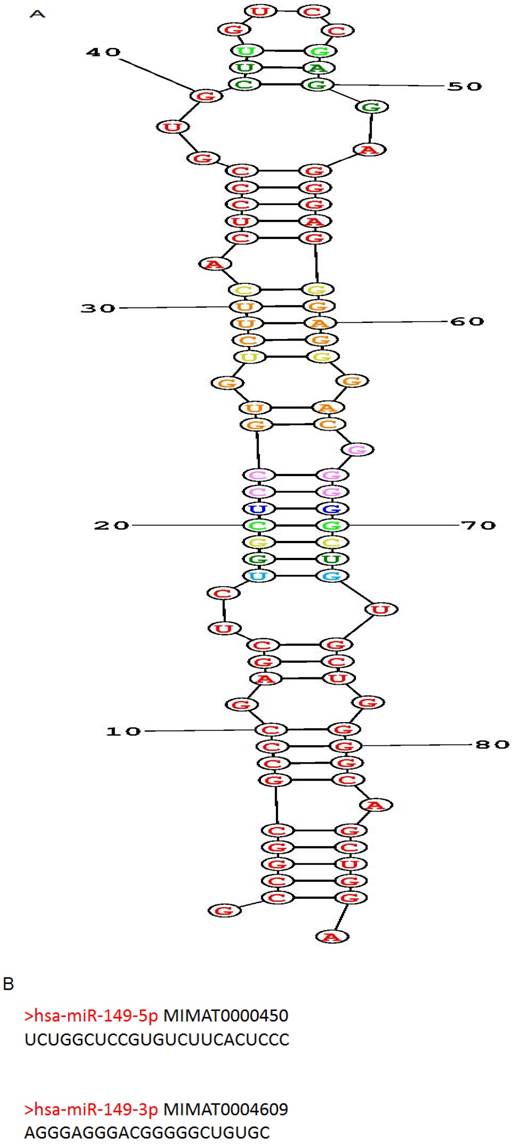
Human miR-149 (hsa-mir-149; MIR149) is located at 2q37.3, encoded by only 1 exon and found to have polymorphisms. The predicted stem-loop structure of miR-149 determined using the RNA structure software is shown in (Figure 1A). The miR-149 hairpin gives rise to the “guide strand” miR-149-5p and the sister "passenger" strand miR-149-3p as shown in (Figure 1B). miR-149-5p is far more prevalent than miR-149-3p according to reads of deep sequencing. And the sequences of miR-149-5p and miR-149-3p are totally dissimilar, indicating inconsistent roles of them involved in human biological behavior.
Technical aspect of miR-149 extraction, detection and confirmation
The researchers initially examined 9 different mouse tissues using tissue-specific cloning and identified 34 novel miRNAs including miR-149, which found expressed higher in mouse heart by northern-blot.3 Then human miR-149 were identified through combined bioinformatics and sequence-directed cloning methods.4 Over 250 small RNA libraries were sequenced from 26 different organ systems and cell types of human and rodents. Detailed and accurate information about mature sequences, precursors, genome location and conservation patterns of miR-149 were provided by this broad research. Moreover, Stephen S.C Chim et al. first detected miR-149 in human blood and confirmed that miR-149 was abundant in maternal plasma during pregnancy.5
Targeted genes and dysregulation of miR-149-5p in various cancers.
| Hsa-miR-149-5p | Cancers | Targeted gene |
|---|---|---|
| Upregulated | ||
| Acute myeloid leukemia | FASLG | |
| Prostate cancer | SOX2, NANOG, Oct4 | |
| Glioblastoma multiforme | Caspase-2 | |
| Metastatic sporadic melanoma | ||
| Downregulated | ||
| Gastric cancer | ZETB2 | |
| Hepatocellular carcinoma | AKT1, PARP-2 | |
| Glioblastoma multiforme | AKT1, Rap1b | |
| Renal cell carcinoma | PPM1F | |
| Lung cancer | FOXM1 | |
| Colorectal cancer | FOXM1, SP1, SRPX2, GPC1, EphB3 | |
| Breast cancer | Rap1a, Rap1b, GIT1, NDST1, ErbB3 | |
| Gastric cancer | IL-6 | |
| Thyroid carcinoma | FOSL1 | |
| Neuroblastoma | Rap1 | |
| Endometrial cancer | ||
| Laryngeal squamous cell carcinoma | ||
| Cervical carcinogenesis | ||
| Astrocytomas |
Targeted genes and dysregulation of miR-149-5p in various cancers.
| Hsa-miR-149-3p | Cancers | Targeted gene |
|---|---|---|
| Upregulated | ||
| Melanoma | GSK3α | |
| T-cell acute lymphoblastic leukemia | JunB | |
| Glioma | ||
| Liver cancer | ||
| Downregulated | ||
| Neuroblastoma | AKT1, E2F1 | |
| Gastric cancer | Wnt-1 | |
| Pancreatic cancer | AKT1 |
The potential interactions of miR-149 with other molecules
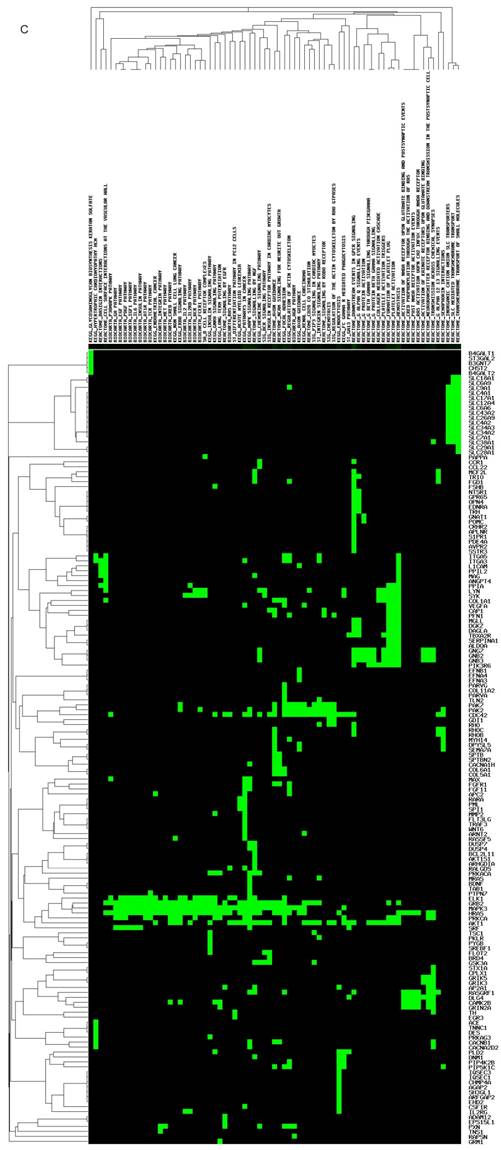
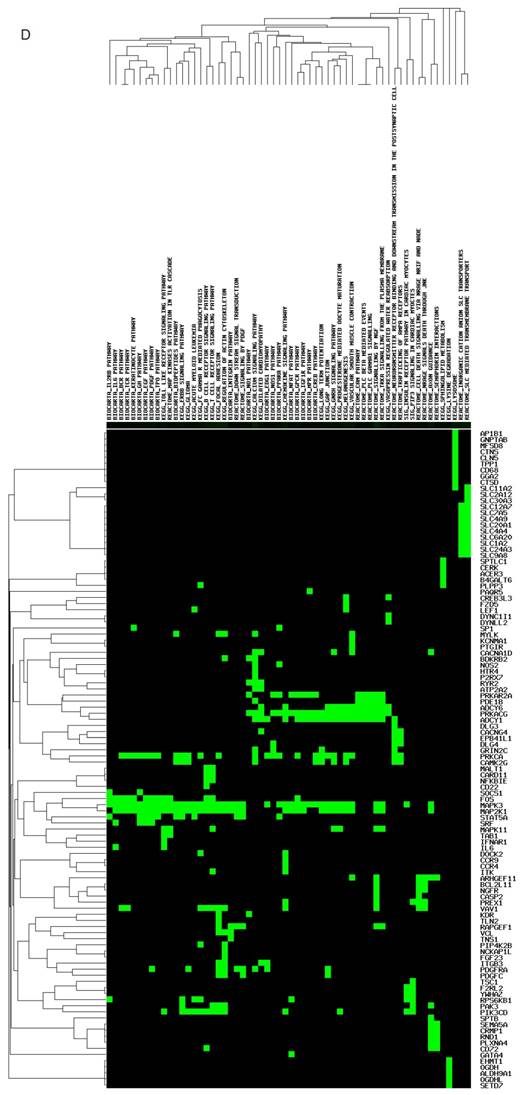
The targeted genes and pathways potentially affected by miR-149-5p and miR-149-3p identified using the Diana-MicroT and TagetScan tools are shown in (Figure 1C, D). Undoubtedly, the gene and pathway signatures of miR-149-5p and miR-149-3p are totally different. B4GAL T1, ST3GAL2, B3GNT7, CHST2, B4GAL T2, SLC18A1, MAG, SYK, VEGFA, PIK3R6, AKT1, RHOC, TNS1 and so on were found as the targeted genes of miR-149-5p. AP1B1, GNPTAB, MFSD8, CTNS, CLN5, TPP1, CD68, IL-6, CCR9, CCR4, PDGFC, NCKAP1L and so forth were predicted as the targeted genes of miR-149-3p.
The potential mechanisms of miR-149's functions
The two isoforms of miR-149 are both predicted to target oncogene and tumor suppressor, leading to dual impacts among cancers. miR-149-5p participates mostly in ERBB-pathway, insulin signaling pathway, MAPK signaling pathway and chemokine signaling pathway, which are necessary for tumor growth, while miR-149-3p plays roles in toll-like receptor signaling pathway, T and B cell receptor signaling pathway, focal adhesion-pathway, vascular smooth muscle contraction-pathway and lysosome-pathway which also related closely to tumorigenesis and tumor progression. In the following sections, we will focus on discussing central roles of miR-149 related to biological functions of tumor, which were confirmed in various experiments and clinical samples.
miR-149 in cancer proliferation and apoptosis
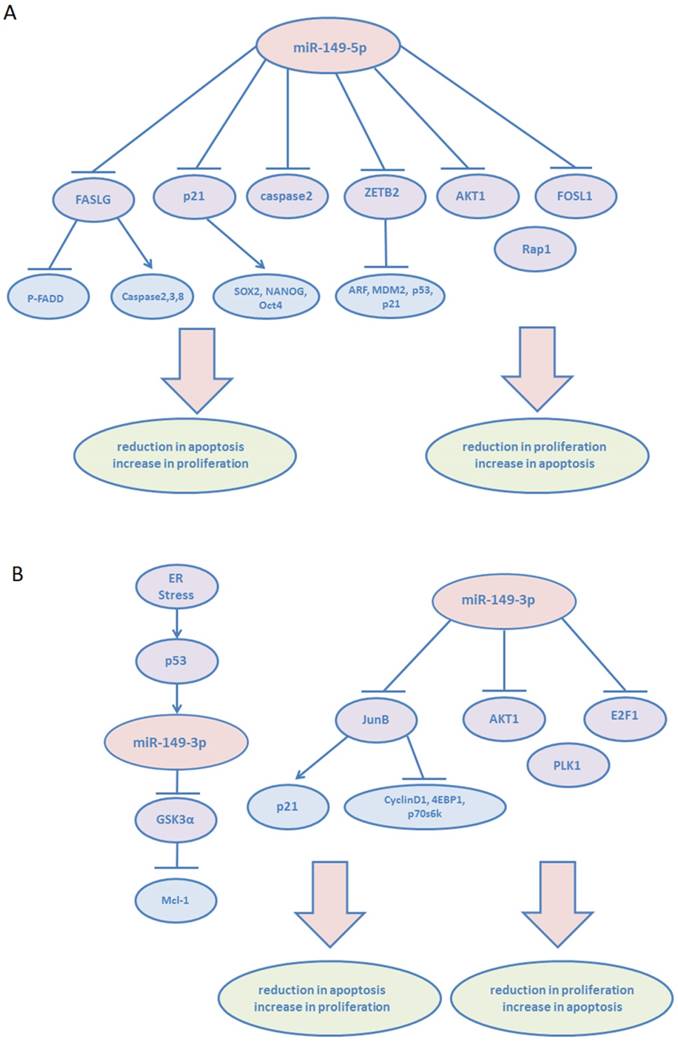
Accelerated proliferation and decreased apoptosis of cancer cells contribute to the progress of tumor development. Tian P et al. described that miR-149-5p suppressed the apoptosis of the acute myeloid leukemia cell line THP-1 by targeting Fas ligand (FASLG), which in turn negatively affected the phosphorylated form of Fas-associated via death domain (p-FADD) and activation of caspases-2, 3, and 8.6 Tomomi Fujii et al demonstrated that decreased miR-149-5p expression level was related to syndecan-1 (CD138, SDC-1), one of the four mammalian heparin sulfate proteoglycans silenced in prostate cancer.7 The increased expression of syndecan-1 contributes to cell survival via regulating NOX-mediated reactive oxygen species (ROS) generation in androgen-independent prostate cancer cells.8 The mRNA levels of the stem cell-related factors, SOX2, NANOG, and Oct4, increased significantly with decrease in miR-149-5p expression. Cell proliferation assay also showed that miR-149-5p promoted cell proliferation by suppressing these factors via p21 reduction. However, in human gastric cancer (GC), miR-149-5p inhibits proliferation and cell cycle progression via targeting ZBTB2, a repressor of the ARF-MDM2-p53-p21 pathway.[9, 10] ZBTB2 has been identified to either repress the transcription of tumor suppressors ARF, p53, and p21, or induce the expression of MDM2 proto-oncogene. Yanqiong Zhang et al investigated the tumor suppressive role of miR-149-5p via targeting AKT1 in human hepatocellular carcinoma (HCC).11 In addition, they identified AKT1 as an unfavorable factor in patients with HCC. miR-149-5p was found to regulate AKT1 expression by integrating bioinformatics and network analysis. The increased expression of miR-149-5p significantly inhibited HCC cell proliferation by regulating AKT1/mTOR pathway. And miR-149-5p inhibited human thyroid carcinoma WRO cells proliferation by targeting FOSL1 according to the study of Hiroki Takeshita et al.12 In human glioblastoma multiforme (GBM), miR-149-5p was also proved to act as a suppressor of AKT1 expression and inhibit the proliferative activities of U251 cell line, which has a mutation in codon 273 of the p53 gene.13 Interestingly, Xiaokun Shen et al showed that the stable overexpression of miR-149-5p leads to the promotion of cell viability of U87MG, a wild-type p53 cell line, inhibition of apoptosis, and induction of tumor xenograft growth in vivo.14 miR-149-5p targets caspase-2 via inactivation of the p53 and p21 pathways. In addition, miR-149-5p may exert opposite biological effects in p53 wild-type and p53 mutants; however, the specific underlying mechanisms remain to be investigated. Furthermore, on the basis of the study of Xu Y et al, miR-149-5p was markedly deregulated in neuroblastoma cell lines and primary tumor tissues associated with poor clinical outcome. In vitro, it was found that miR-149-5p downregulation promoted proliferation of neuroblastoma cells via targeting the small GTPases: Rap1.15
Similar to miR-149-5p, miR-149-3p plays dual roles in cancer. Unlike other cancers, in human melanoma, wild-type p53 is often expressed at high levels.16 Lei Jin et al detected upregulated miR-149-3p expression in melanoma cell lines in response to endoplasmic reticulum (ER) stress.17 Next, they demonstrated that under ER stress, p53 binding to the p53-binding region at the GPC1 gene transcriptionally upregulates miR-149-3p expression. Furthermore, miR-149-3p targets the 3′UTR of GSK3α, which is known to target Mcl-1, consequently leading to apoptosis resistance in melanoma cells.18 miR-149-3p overexpression was observed in fresh human metastatic melanoma isolates. miR-149-3p-mediated pathway provided a reasonable explanation for the ineffectiveness of wild-type p53 in suppressing melanoma. miR-149-3p was also shown to serve as an oncogenic regulator by promoting proliferation and reducing apoptosis in T-cell acute lymphoblastic leukemia (T-ALL) cells via targeting JunB, and subsequently downregulating p21 as well as upregulating cyclin D1, 4EBP1, and p70s6k.19 However, miR-149-3p was found to suppress the expression of polo-like kinase 1 (PLK1), a critical regulator of cell cycle progression and apoptosis, by targeting the 3'UTR of its mRNA, leading to tumor cell clonogenicity decrease and apoptosis induction.20 In addition, in human miR-149-3p-transfected neuroblastoma and HeLa cell lines, caspase3,7 activity was induced and cell growth was inhibited significantly.21 AKT1 and E2F1 were proven to be the direct targets of miR-149-3p. Importantly, an inverse correlation of miR-149-3p with E2F1 expression (P=0.026) was revealed in primary neuroblastoma samples. Furthermore, miR-149-3p was found upregulated in U87-MG cells following treatment of quinidine, a voltage-gated K+ channel blocker, leading to proliferation inhibition and apoptosis induction of glioma cells.22
miR-149 in cancer migration and invasion
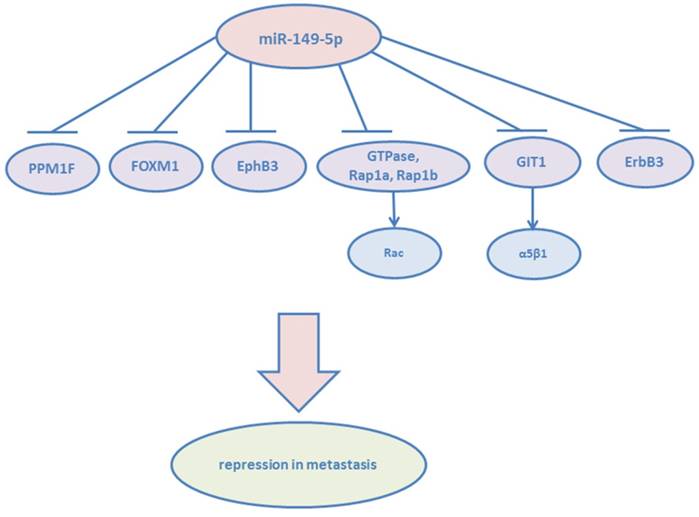
Cancer metastasis accounts for almost all cancer-related deaths. Metastasis is a multistep process, involving tumor cell dissemination from the primary tumor and colonizing distant organs.23 The improved ability of tumor cells to undergo migration and invasion contributes to the first step of this process. miR-149-5p was significantly downregulated in RCC tissues. The overexpression of miR-149-5p contributed to the suppression of cancer cell proliferation and migration.24 Forkhead box transcription factor (FOXM1) is a typical transcription factor and defined as having a common DNA-binding domain called Forkhead or winged-helix domain.25 FOXM1 regulates both G1-S and G2-M phases of cell cycle and mitotic spindle integrity, and its overexpression leads to tumor angiogenesis and metastasis.26 And in colorectal cancer, miR-149-5p suppresses cell migration and invasion by targeting FOXM1,27 in non-small cell lung cancer, epithelial-to-mesenchymal (EMT) of cancer cells are inhibited by miR-149-5p via targeting FOXM1.28 Luo G et al demonstrated that miR-149-5p repressed the metastasis of hepatocellular cells in vitro and in vivo by targeting the actin-regulatory proteins PPM1F.29 PPM1F can promote the formation of stress fibers, which play an important role in metastasis.30 Moreover, miR-149-5p was found able to suppress migration and invasion of human colonic carcinoma HCT116 and SW620 cell lines by targeting an oncogene: EphB3.31 The study by Bischoff A et al showed that miR-149-5p functioned as a tumor suppressor via impairing cell spreading, migration, and invasion of basal-like breast cancer cells via targeting the small GTPases :Rap1a, Rap1b and downstream of integrin receptors. Cell metastasis depends on the activation of Rac, which is known to drive mesenchymal type of cancer cell motility.32 miR-149-5p has been proven to reduce Rac activation indirectly by Rap1a/b co-depletion. They also verified miR-149-5p was able to block the growth factor heregulin (HRG) and PI3K-Akt passway via targeting ErbB3, whose deregulated expression contributes to the transformed phenotype of breast cancer cells.33 Moreover, cancer cells transfected with miR-149-5p blocked lung colonization in vivo. miR-149-5p targets G-protein-coupled receptor kinase-interacting protein (GIT1), another member of integrin signaling.34 It has been known that GIT1 plays a central role in regulating fibronectin-induced focal adhesion formation and cell migration.[35, 36] Overexpression of miR-149-5p reduced the phosphorylation of focal adhesion kinase and paxillin, but enhanced the degradation of paxillin and α5β1 integrin. GIT1 depletion reduced α5β1-integrin-mediated cell adhesion to fibronectin. High GIT1 level and low miR-149-5p level were significantly associated with the advanced stages of breast cancer, especially lymph node metastasis.
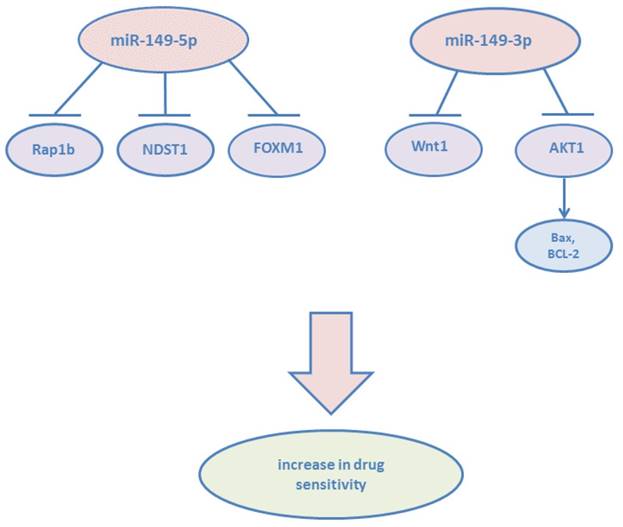
miR-149 in drug sensitivity and resistance
Cancer resistance to chemotherapy, molecularly targeted therapies, and other anti-cancer drugs limits treatment effectiveness.37 Studies have shown that miR-149 participates in regulating drug sensitivity and cancer resistance. In GBM, miR-149-5p was downregulated, while the chemosensitivity of glioblastoma cells to temozolomide (TMZ) was enhanced via targeting Rap1b.38 Recent years, studies have shown that one type of dietary restrictions (DR): short time starvation (STS) is able to make chemotherapy more suitable.39 Higher expression level of miR-149-5p in breast cancer cells: MDA-MB-231 and normal mammary epithelial cells: MCF10A treated with doxorubicin under STS for 48h could explain that miR-149-5p was involved in pathways related to increase drug sensitivity.40 In adriamycin (ADM)-resistant human breast cancer cells, He DX et al demonstrated that the downregulation of miR-149-5p was attributed to hypermethylation of its 5'-UTR.41 Chemoresistance was suppressed by miR-149-5p through targeting GlcNAc N-deacetylase/N-sulfotransferase-1(NDST1). NDST1 has the ability to activate a heparin sulfate-related pathway involving heparanase, which contributes to chemoresistance. miR-149-5p not only decreases cell migration and invasion, but also increases the sensitivity of colorectal cancer cells to 5-Fluorouracil (5-FU) via targeting FOXM1.42 In 5-FU-responding colorectal cancer tissues, the expression of miR-149-5p was significantly higher than that in non-responding tissues and inversely correlated with the mRNA level of FOXM1. It has been found that high level of bisphenol A (BPA) is one of risk factors to induce endometrial cancer (EC). The expression of miR-149-5p was reduced after BPA exposure, then leads to down-regulate DNA repair gene ADP-ribosylation factor 6 (ARF6) and tumor protein p53 (TP53).43 These finding revealed a potential epigenetic mechanism among the risk of endometrial carcinogenesis. However, miR-149-5p was found upregulated in gefitibib-resistant human lung adenocarcinoma cell line and involved in the acquired gefitinib resistance.44
The study by Donghui Cao et al. revealed that 18β-glycyrrhetinic acid (GRA) could attenuate the severity of gastritis, suppress gastric tumorigenesis in transgenic mice, ameliorate the inflammatory microenvironment through the inhibition of COX-2 expression, and upregulate the expression of miR-149-3p which targeted Wnt-1.45 In addition, dioscin, a natural product, is an active ingredient in some medicinal plants and has shown anti-tumor effects against many cancers.46 In pancreatic cancer, dioscin was found to induce apoptosis and suppress the tumor growth of ASPC-1 and PANC-1 cell xenografts by upregulating miR-149-3p expression which targeted AKT1, increasing the expression levels of Bax, Apaf-1, cleaved caspase-3/9, and cleaved PARP, and suppressing Bcl-2 levels, which results in cytochrome c release.47 Bafilomycin A1 is an inhibitor of the c subunit of the V-ATPase and has been investigated able to inhibit the proliferation and metastasis of cancer cells.48 miR-149-3p was also found upregulated in BEL-7402 liver cancer and HO-8910 ovarian cancer cell lines following bafilomycin A1 treatment.49 However, on the basis of the study of Ma J et al, microcystin-LR (MC-LR) has been identified as one of risk factors for human primary hepatocellular cancer.[50, 51] Exposure of MC-LR was found promoted the expression of miR-149-3p, promoting miR-149-3p may be involved in MC-LR-hepatotoxicity, hepatitis and liver carcinoma.52
miR-149 in tumor microenvironment
Tumor microenvironment is now recognized as an important participant in metastatic progression and therapeutic responses.53 Extracellular matrix (ECM) and numerous stromal cells, including blood endothelial cells, lymphatic endothelial cells, cancer-associated fibroblasts (CAFs), tumor-associated macrophages (TAMs), mesenchymal stem cells (MSCs), immune cells, leukocytes, and adipocytes, constitute the tumor microenvironment.[54, 55]
Structure, targeted genes and pathways of miR-149. (A): The predicted stem-loop structure of miR-149 determined using the RNA structure software. (B): The sequence of miR-149. (C, D): The targeted genes and pathways potentially affected by miR-149-5p and miR-149-3p identified using the Diana-MicroT and TagetScan tools.
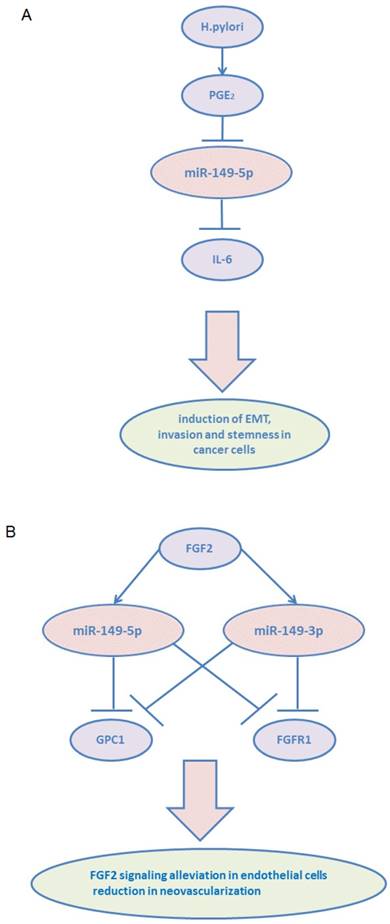
Among the stromal cells, heterogeneous populations of CAFs are key players in multicellular stromal-dependent alterations that contribute to malignant initiation and progression.56 In addition, crosstalk between CAFs and tumor cells lead to tumor progression. CAFs are usually considered to be derived from normal fibroblasts (NFs), and they communicate with tumor cells via growth and inflammation factors.[57, 58] Inflammatory signaling pathways are considered highly promising targets to block the crosstalk between tumor cells and CAFs.59 Pu Li et al demonstrated that miR-149-5p links PGE2 and IL-6 signaling in mediating the communication between tumor cells and CAFs in GC.60 They showed that miR-149-5p regulated fibroblast activation protein (FAP) expression via targeting IL-6, and its expression was downregulated in GC CAFs. miR-149-5p was found to inhibit fibroblast activation by reducing IL-6 expression. In addition, CAFs enhanced epithelial-to-mesenchymal transition and the stem-like properties of GC cells in a miR-149-IL-6-dependent manner. Besides IL-6, PGE2 receptor 2 (PTGER2/EP2) was revealed as another target of miR-149-5p. H.pylori infection, a leading cause of human GC, caused the hypermethylation of miR-149-5p and induced COX-2/PGE2 as well as IL-6 signaling pathways.
Fibroblast growth factors (FGFs) and their receptors (FGFRs) regulate numerous cellular processes, especially in the endothelial cells.61 Glypicans (GPCs) are frequently overexpressed in many cancers and recognized to play a role in tumorigenesis and angiogenesis.62 GPC1 promoted FGF2 binding to its receptor (FGFR1), subsequently enhancing FGF2-FGFR1 activation and signaling. miR-149, including miR-149-5p and miR-149-3p, was found to be located within the first intron of GPC1 gene and showed targeting activity towards both GPC1 and FGFR1 in endothelial cells.63 Decreased FGF2 signaling in endothelial cells and reduced neovascularization were observed in vivo following miR-149 overexpression. Notably, FGF2 was found to stimulate the expression of miR-149, thereby assuring the steady state of FGF2-induced responses in endothelial cells.
miR-149 as potential therapeutic targets
As we discussed above, miR-149 plays important roles in tumorigenesis and tumor progression of various cancers. There also have been a large number of studies about exploring the effect using miR-149 as therapeutic targets. Chen Y et al observed a reduction in proliferation and increase in the apoptosis of prostate cancer stem cells from TSU cell line via the downregulated expression of miR-149-5p.64 Proliferation inhibition and apoptosis induction were observed using expression enhancement of miR-149-3p in glioma cells.22 Moreover, among various cancers, metastasis was repressed using transfecting miR-149-5p mimics in vitro and in vivo.[29, 35, 36] And chemoresistance to 5-Fluorouracil was suppressed by introducing miR-149-5p mimics in colorectal cancer cells.42 Furthermore, fibroblast activation inhibition and neovascularization reduction were achieved by increasing expression of miR-149-5p in CAFs and endothelial cells.[60, 63]
miR-149 as potential biomarkers
Numerous considerable efforts have been devoted to identifying suitable miRNAs as diagnostic, prognostic markers, owing to their remarkably stable forms and abnormal expression in cancer patients compared to that in healthy individuals.65 Single-nucleotide polymorphisms (SNPs) in microRNA represent another type of genetic variability that can influence the risk and outcome of human disease.66 In Rong GQ, Dikeakos P and Li L et al's study, miR-149-5p gene polymorphisms was found associated with susceptibility to CRC and GC,[67-69] particularly in males.70 In Pan XM, Liu XX and Du W et al's study, they also revealed that single nucleotide (SNPs) rs2292832 in miR-149-5p was related to high CRC susceptibility.[71, 72] Jia H , Wang XH and Feng Y et al respectively confirmed that rs2292832 in miR-149-5p contributed to the risk of hepatocellular cancer and breast cancer,[73-75] especially in HBV-infected HCC patients.76 PS and Wei WJ et al also found miR-149-5p rs2292832 C>T contributed to the development of oral squamous cell carcinoma in South Indian Population and the risk of papillary thyroid cancer in Chinese population.[77, 78] In addition, pre-miR-149-5p rs71428439 polymorphism was investigated associated with increased cancer risk in hepatocellular carcinoma and head and neck squamous cell carcinoma. The mechanism of this SNP possibly involved downregulated miR-149-5p expression and upregulation targeted gene AKT1 expression.[79, 80] The study from Wang Z et al also demonstrated that the GG genotypes of miR-149-5p rs71428439 increased the risk of clear cell renal cell carcinoma.81 But miR-149-5p rs2292832 T/C polymorphism could decrease digestive cancer susceptibility in Li L et al's meta-analysis study.82 Wang et al found that pre-mir-149-5p rs2292382 had significantly better survival in laryngeal squamous cell carcinoma.83 Huang GL et al demonstrated miR-149-5p rs2292832 contributed to the progression rather than the initiation of nasopharyngeal cancer.84 Furthermore, miR-149-5p C-containing genotypes of miR-149-5p rs2292832 was verified associated with better over survival (OS), and might function through DNA topoisomerases 1 (TOP1) pathway in non-small cell lung cancer.[85, 86] And in colorectal cancer, miR-149-5p was found to be epigenetically silenced, which was associated with the hypermethylation of the neighboring CpG Island. miR-149-5p acts as a tumor suppressor via targeting a potential oncogenic protein, Specificity Protein 1 (SP1).[87, 88] Wilting SM et al also demonstrated methylation-mediated transcriptional repression of miR-149-5p participated in human papillpmavirus (HPV) induced cervical carcinogenesis.89 Another study demonstrated that miR-149-5p was downregulated in colorectal cancer, and it regulated the expression of SRPX2.90 AKT1 was identified as an unfavorable prognostic factor for patients with HCC. Zhang Y et al found that miR-149 functions as a tumor suppressive miRNA via targeting AKT1 in patients with HCC by using five online programs for miRNA target prediction.11 Lin L et al studied the expression of miR-149-5p and PARP-2 in HCC tumor tissues, and their roles in sensitizing chemo/radiotherapy. The high expression level of PARP-2 and low expression level of miR-149-5p were found to be correlated with larger tumor mass, increased capsular and vascular invasion, and lymph node metastases. High PARP-2 and low miR-149-5p levels were identified as poor prognosis factors, and they reduced sensitivity to chemotherapy and radiotherapy in xenogratft HCC animal models.91 Xue L et al aimed to investigate the prognostic value of miR-149-5p in glioma.92 They found that the expression of miR-149-5p was significantly lower in tumor tissues compared to that in normal tissues. Improved survival duration and lower mortality risk were also observed using Kaplan-Meier and multivariate analysis. Furthermore, it was revealed that AKt/mTOR signaling was hyperactive in low miR-149 expressing tissues. miR-149-5p was also proven to play critical roles in astrocytomas and laryngeal squamous cell carcinoma.[93, 94] miR-149-5p expression was significantly lower in tumor tissues than in normal brain and vocal cord polyp tissues. Shorter survival duration was seen in patients with lower miR-149-5p expression. However, in Yang C et al' s study, the expression of miR-149-5p was reported to be increased in NSCLC tissues using either small RNA-sequencing or qRT-PCR validation.95 miR-149-5p was also found overexpressed in castration-resistant prostate cancer. And in upper tract urothelial carcinoma, miR-149-5p was identified as independently associated with cancer-specific survival.96 Besides, overexpression of miR-149-3p was found in plasma of melanoma patients compared with healthy controls. miR-149-3p was identified as a biomarker with high diagnostic power in early stage of melanoma.97 And miR-149-3p overexpression was also investigated strongly correlated in uveal melanoma patients with liver metastasis.98
Recently, serum or plasma exosomal miRNAs have emerged as novel markers for diagnosis and prognostic evaluation in patients with various cancers.99 Extracellular vesicles (EVs) and exosomes are membrane-derived vesicles that are actively secreted by cells. EVs with a diverse range of sizes (100-1,000 nm in diameter) contain microvesicles and ectosomes, which originate by budding from the plasma membrane of cells. Exosomes, typically smaller than 150 nm in diameter, are generated inside multivesicular endosomes or multivesicular bodies (MVBs). When these compartments fuse with the plasma membrane, exosome secretion is activated.100 A previous study showed that EVs and exosomes contain numerous molecules, such as cell-specific proteins, lipids, mRNAs, and abundant miRNAs.[101-103] Owing to the protective function of EVs or exosomes against degradation by RNAse enzymes, exosomal miRNAs are stable in blood or other body fluids, making them ideal candidates for biomarkers in cancers. Pfeffer SR et al. showed that exosomal miR-149-5p was expressed at significantly higher levels in patients with metastatic sporadic melanoma compared to that in familial melanoma patients or unaffected control subjects, suggesting that miR-149-5p could be a poor prognostic factor in melanoma.104 As we mentioned above, miR-149-5p was found to have targeting activity towards GPC1 in endothelial cells. Similarly, miR-149-5p decreased GPC1 expression in colorectal cancer cell lines. Moreover, miR-149-5p was significantly decreased in GPC+ exosomes from tumor tissues and plasma of colorectal cancer patients compared to that in peritumoural tissues and plasma of healthy controls.105
Conclusions
miR-149-5p or miR-149-3p plays dual roles in the proliferation and apoptosis of various tumors (Figure 2). In cancer metastasis, miR-149-5p is found to be an important tumor suppressor, and its expression was downregulated (Figure 3). Currently, both miR-149-5p and miR-149-3p are confirmed to increase drug sensitivity according to most of the literature (Figure 4). In tumor microenvironment, miR-149-5p regulated FAP expression on CAFs via targeting IL-6, leading to fibroblast activation inhibition and its expression was downregulated in gastric cancer. Overexpression of miR-149-5p or miR-149-3p alleviated FGF2 signaling in endothelial cells and reduced neovascularization (Figure 5). miR-149-5p gene polymorphisms contribute to alter susceptibility to cancers. Therefore, it can be suggested that miR-149 could serve as biomarkers and therapeutic targets in numerous cancers.
Dual roles of miR-149-5p and miR-149-3p in proliferation and apoptosis. (A): miR-149-5p. (B): miR-149-3p.
miR-149-5p found to be an important tumor suppressor of metastasis.
miR-149-5p and miR-149-3p confirmed to increase drug sensitivity.
miR-149-3p and miR-149-5p participate in tumor microenvironment. (A) miR-149-5p mediates the crosstalk between tumor cells and CAFs: H. pylori infection leads to induce PGE2 signaling and result in reduction of miR-149-5p in CAFs and increase IL-6 secretion. (B) Overexpression of miR-149-5p or miR-149-3p alleviated FGF2 signaling in endothelial cells and reduced neovascularization via targeting GPC1 and FGFR1.
Acknowledgements
This research was supported by National Program on Key Research Project (No. 2016YFC0905900), Medical Training Programme Foundation for the Talents by Jiangsu Provincial Department of health (No.17 [2016]), The 333 Talent Project of Jiangsu Province, top-level (No.4 [2016]), the National key clinical specialist construction Programs of China (No.544 [2013]), Major Program of Natural Science Foundation of Jiangsu Province (No. BL2014090), Natural Science Foundation of Jiangsu Province (No. BK20151579), A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD, JX10231802). This study was also funded by the National Natural Science Foundation of China (grant number 81602551), the Key Research Program for the Social Development of Jiangsu Province, China (BE2015718) and the National Natural Science Foundation of China (No. 81602551).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nature cell biology. 2009;11:228-34
2. Melo SA, Esteller M. Dysregulation of microRNAs in cancer: playing with fire. FEBS letters. 2011;585:2087-99
3. Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Current biology: CB. 2002;12:735-9
4. Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A. et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401-14
5. Chim SSC, Shing TKF, Hung ECW, Leung Ty, Lau Tk, Chiu RWK. et al. Detection and Characterization of Placental MicroRNAs in Maternal Plasma. Clinical Chemistry. 2008;54:482-90
6. Tian P, Yan L. Inhibition of MicroRNA-149-5p Induces Apoptosis of Acute Myeloid Leukemia Cell Line THP-1 by Targeting Fas Ligand (FASLG). Medical science monitor: international medical journal of experimental and clinical research. 2016;22:5116-23
7. Fujii T, Shimada K, Tatsumi Y, Fujimoto K, Konishi N. Syndecan-1 responsive microRNA-126 and 149 regulate cell proliferation in prostate cancer. Biochem Biophys Res Commun. 2015;456:183-9
8. Shimada K, Nakamura M, De Velasco MA, Tanaka M, Ouji Y, Konishi N. Syndecan-1, a new target molecule involved in progression of androgen-independent prostate cancer. Cancer Sci. 2009;100:1248-54
9. Wang Y, Zheng X, Zhang Z, Zhou J, Zhao G, Yang J. et al. MicroRNA-149 inhibits proliferation and cell cycle progression through the targeting of ZBTB2 in human gastric cancer. PLoS One. 2012;7:e41693
10. Jeon BN, Choi WI, Yu MY, Yoon AR, Kim MH, Yun CO. et al. ZBTB2, a novel master regulator of the p53 pathway. The Journal of biological chemistry. 2009;284:17935-46
11. Zhang Y, Guo X, Xiong L, Yu L, Li Z, Guo Q. et al. Comprehensive analysis of microRNA-regulated protein interaction network reveals the tumor suppressive role of microRNA-149 in human hepatocellular carcinoma via targeting AKT-mTOR pathway. Molecular cancer. 2014;13:253
12. Takeshita H, Shiozaki A, Bai XH, Iitaka D, Kim H, Yang BB. et al. XB130, a new adaptor protein, regulates expression of tumor suppressive microRNAs in cancer cells. PLoS One. 2013;8:e59057
13. Pan SJ, Zhan SK, Pei BG, Sun QF, Bian LG, Sun BM. MicroRNA-149 inhibits proliferation and invasion of glioma cells via blockade of AKT1 signaling. International journal of immunopathology and pharmacology. 2012;25:871-81
14. Shen X, Li J, Liao W, Wang J, Chen H, Yao Y. et al. microRNA-149 targets caspase-2 in glioma progression. Oncotarget. 2016;7:26388-99
15. Xu Y, Chen X, Lin L, Chen H, Yu S, Li D. MicroRNA-149 is associated with clinical outcome in human neuroblastoma and modulates cancer cell proliferation through Rap1 independent of MYCN amplification. Biochimie. 2017;139:1-8
16. Sparrow LE, Soong R, Dawkins HJ, Iacopetta BJ, Heenan PJ. p53 gene mutation and expression in naevi and melanomas. Melanoma research. 1995;5:93-100
17. Jin L, Hu WL, Jiang CC, Wang JX, Han CC, Chu P. et al. MicroRNA-149*, a p53-responsive microRNA, functions as an oncogenic regulator in human melanoma. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:15840-5
18. Maurer U, Charvet C, Wagman AS, Dejardin E, Green DR. Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Molecular cell. 2006;21:749-60
19. Fan SJ, Li HB, Cui G, Kong XL, Sun LL, Zhao YQ. et al. miRNA-149* promotes cell proliferation and suppresses apoptosis by mediating JunB in T-cell acute lymphoblastic leukemia. Leukemia research. 2016;41:62-70
20. Shin CH, Lee H, Kim HR, Choi KH, Joung JG, Kim HH. Regulation of PLK1 through competition between hnRNPK, miR-149-3p and miR-193b-5p. Cell Death Differ. 2017
21. Lin RJ, Lin YC, Yu AL. miR-149* induces apoptosis by inhibiting Akt1 and E2F1 in human cancer cells. Molecular carcinogenesis. 2010;49:719-27
22. Ru Q, Tian X, Pi MS, Chen L, Yue K, Xiong Q. et al. Voltagegated K+ channel blocker quinidine inhibits proliferation and induces apoptosis by regulating expression of microRNAs in human glioma U87MG cells. International journal of oncology. 2015;46:833-40
23. Turajlic S, Swanton C. Metastasis as an evolutionary process. Science (New York, NY). 2016;352:169-75
24. Jin L, Li Y, Liu J, Yang S, Gui Y, Mao X. et al. Tumor suppressor miR-149-5p is associated with cellular migration, proliferation and apoptosis in renal cell carcinoma. Mol Med Rep. 2016;13:5386-92
25. Katoh M, Katoh M. Human FOX gene family (Review). International journal of oncology. 2004;25:1495-500
26. Halasi M, Gartel AL. Targeting FOXM1 in cancer. Biochemical pharmacology. 2013;85:644-52
27. Xu K, Liu X, Mao X, Xue L, Wang R, Chen L. et al. MicroRNA-149 suppresses colorectal cancer cell migration and invasion by directly targeting forkhead box transcription factor FOXM1. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2015;35:499-515
28. Ke Y, Zhao W, Xiong J, Cao R. miR-149 Inhibits Non-Small-Cell Lung Cancer Cells EMT by Targeting FOXM1. Biochemistry research international. 2013;2013:506731
29. Luo G, Chao YL, Tang B, Li BS, Xiao YF, Xie R. et al. miR-149 represses metastasis of hepatocellular carcinoma by targeting actin-regulatory proteins PPM1F. Oncotarget. 2015;6:37808-23
30. Phang HQ, Hoon JL, Lai SK, Zeng Y, Chiam KH, Li HY. et al. POPX2 phosphatase regulates the KIF3 kinesin motor complex. Journal of cell science. 2014;127:727-39
31. Zhang G, Liu X, Li Y, Wang Y, Liang H, Li K. et al. EphB3-targeted regulation of miR-149 in the migration and invasion of human colonic carcinoma HCT116 and SW620 cells. Cancer Sci. 2017;108:408-18
32. Bischoff A, Huck B, Keller B, Strotbek M, Schmid S, Boerries M. et al. miR149 functions as a tumor suppressor by controlling breast epithelial cell migration and invasion. Cancer Res. 2014;74:5256-65
33. Bischoff A, Bayerlova M, Strotbek M, Schmid S, Beissbarth T, Olayioye MA. A global microRNA screen identifies regulators of the ErbB receptor signaling network. Cell communication and signaling: CCS. 2015;13:5
34. Chan SH, Huang WC, Chang JW, Chang KJ, Kuo WH, Wang MY. et al. MicroRNA-149 targets GIT1 to suppress integrin signaling and breast cancer metastasis. Oncogene. 2014;33:4496-507
35. Manabe R, Kovalenko M, Webb DJ, Horwitz AR. GIT1 functions in a motile, multi-molecular signaling complex that regulates protrusive activity and cell migration. Journal of cell science. 2002;115:1497-510
36. Schmalzigaug R, Garron ML, Roseman JT, Xing Y, Davidson CE, Arold ST. et al. GIT1 utilizes a focal adhesion targeting-homology domain to bind paxillin. Cellular signalling. 2007;19:1733-44
37. Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nature reviews Cancer. 2013;13:714-26
38. She X, Yu Z, Cui Y, Lei Q, Wang Z, Xu G. et al. miR-128 and miR-149 enhance the chemosensitivity of temozolomide by Rap1B-mediated cytoskeletal remodeling in glioblastoma. Oncol Rep. 2014;32:957-64
39. Raffaghello L, Lee C, Safdie FM, Wei M, Madia F, Bianchi G. et al. Starvation-dependent differential stress resistance protects normal but not cancer cells against high-dose chemotherapy. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:8215-20
40. Rizzo S, Cangemi A, Galvano A, Fanale D, Buscemi S, Ciaccio M. et al. Analysis of miRNA expression profile induced by short term starvation in breast cancer cells treated with doxorubicin. Oncotarget. 2017
41. He DX, Gu XT, Li YR, Jiang L, Jin J, Ma X. Methylation-regulated miR-149 modulates chemoresistance by targeting GlcNAc N-deacetylase/N-sulfotransferase-1 in human breast cancer. The FEBS journal. 2014;281:4718-30
42. Liu X, Xie T, Mao X, Xue L, Chu X, Chen L. MicroRNA-149 Increases the Sensitivity of Colorectal Cancer Cells to 5-Fluorouracil by Targeting Forkhead Box Transcription Factor FOXM1. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2016;39:617-29
43. Chou WC, Lee PH, Tan YY, Lin HC, Yang CW, Chen KH. et al. An integrative transcriptomic analysis reveals bisphenol A exposure-induced dysregulation of microRNA expression in human endometrial cells. Toxicology in vitro: an international journal published in association with BIBRA. 2017;41:133-42
44. Hu Y, Qin X, Yan D, Cao H, Zhou L, Fan F. et al. Genome-wide profiling of micro-RNA expression in gefitinib-resistant human lung adenocarcinoma using microarray for the identification of miR-149-5p modulation. Tumour Biol. 2017;39:1010428317691659
45. Cao D, Jia Z, You L, Wu Y, Hou Z, Suo Y. et al. 18beta-glycyrrhetinic acid suppresses gastric cancer by activation of miR-149-3p-Wnt-1 signaling. Oncotarget. 2016;7:71960-73
46. Lu B, Xu Y, Xu L, Cong X, Yin L, Li H. et al. Mechanism investigation of dioscin against CCl4-induced acute liver damage in mice. Environmental toxicology and pharmacology. 2012;34:127-35
47. Si L, Xu L, Yin L, Qi Y, Han X, Xu Y. et al. Potent effects of dioscin against pancreatic cancer via miR-149-3P-mediated inhibition of the Akt1 signalling pathway. British journal of pharmacology. 2017;174:553-68
48. Morimura T, Fujita K, Akita M, Nagashima M, Satomi A. The proton pump inhibitor inhibits cell growth and induces apoptosis in human hepatoblastoma. Pediatric surgery international. 2008;24:1087-94
49. Lu X, Chen L, Chen Y, Shao Q, Qin W. Bafilomycin A1 inhibits the growth and metastatic potential of the BEL-7402 liver cancer and HO-8910 ovarian cancer cell lines and induces alterations in their microRNA expression. Exp Ther Med. 2015;10:1829-34
50. Ueno Y, Nagata S, Tsutsumi T, Hasegawa A, Watanabe MF, Park HD. et al. Detection of microcystins, a blue-green algal hepatotoxin, in drinking water sampled in Haimen and Fusui, endemic areas of primary liver cancer in China, by highly sensitive immunoassay. Carcinogenesis. 1996;17:1317-21
51. Chen JG, Kensler TW. Changing rates for liver and lung cancers in Qidong, China. Chemical research in toxicology. 2014;27:3-6
52. Ma J, Li Y, Yao L, Li X. Analysis of MicroRNA Expression Profiling Involved in MC-LR-Induced Cytotoxicity by High-Throughput Sequencing. Toxins. 2017:9
53. Place AE, Jin Huh S, Polyak K. The microenvironment in breast cancer progression: biology and implications for treatment. Breast cancer research: BCR. 2011;13:227
54. Turley SJ, Cremasco V, Astarita JL. Immunological hallmarks of stromal cells in the tumour microenvironment. Nature reviews Immunology. 2015;15:669-82
55. Mao Y, Keller ET, Garfield DH, Shen K, Wang J. Stromal cells in tumor microenvironment and breast cancer. Cancer metastasis reviews. 2013;32:303-15
56. Gascard P, Tlsty TD. Carcinoma-associated fibroblasts: orchestrating the composition of malignancy. Genes & development. 2016;30:1002-19
57. Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R. et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335-48
58. Swartz MA, Iida N, Roberts EW, Sangaletti S, Wong MH, Yull FE. et al. Tumor microenvironment complexity: emerging roles in cancer therapy. Cancer Res. 2012;72:2473-80
59. Albini A, Sporn MB. The tumour microenvironment as a target for chemoprevention. Nature reviews Cancer. 2007;7:139-47
60. Li P, Shan JX, Chen XH, Zhang D, Su LP, Huang XY. et al. Epigenetic silencing of microRNA-149 in cancer-associated fibroblasts mediates prostaglandin E2/interleukin-6 signaling in the tumor microenvironment. Cell Res. 2015;25:588-603
61. Babina IS, Turner NC. Advances and challenges in targeting FGFR signalling in cancer. Nature reviews Cancer. 2017;17:318-32
62. Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nature reviews Cancer. 2015;15:540-55
63. Chamorro-Jorganes A, Araldi E, Rotllan N, Cirera-Salinas D, Suarez Y. Autoregulation of glypican-1 by intronic microRNA-149 fine tunes the angiogenic response to FGF2 in human endothelial cells. Journal of cell science. 2014;127:1169-78
64. Chen Y, Zhao J, Luo Y, Wang Y, Jiang Y. Downregulated expression of miRNA-149 promotes apoptosis in side population cells sorted from the TSU prostate cancer cell line. Oncol Rep. 2016;36:2587-600
65. Cheng G. Circulating miRNAs: roles in cancer diagnosis, prognosis and therapy. Advanced drug delivery reviews. 2015;81:75-93
66. Chen K, Song F, Calin GA, Wei Q, Hao X, Zhang W. Polymorphisms in microRNA targets: a gold mine for molecular epidemiology. Carcinogenesis. 2008;29:1306-11
67. Rong GQ, Zhang XM, Chen B, Yang XD, Wu HR, Gong W. MicroRNA gene polymorphisms and the risk of colorectal cancer. Oncol Lett. 2017;13:3617-23
68. Dikeakos P, Theodoropoulos G, Rizos S, Tzanakis N, Zografos G, Gazouli M. Association of the miR-146aC>G, miR-149T>C, and miR-196a2T>C polymorphisms with gastric cancer risk and survival in the Greek population. Mol Biol Rep. 2014;41:1075-80
69. Li L, Sheng Y, Lv L, Gao J. The association between two microRNA variants (miR-499, miR-149) and gastrointestinal cancer risk: a meta-analysis. PLoS One. 2013;8:e81967
70. Zhang MW, Jin MJ, Yu YX, Zhang SC, Liu B, Jiang X. et al. Associations of lifestyle-related factors, hsa-miR-149 and hsa-miR-605 gene polymorphisms with gastrointestinal cancer risk. Molecular carcinogenesis. 2012;51(Suppl 1):E21-31
71. Pan XM, Xiao X, Qin HJ, Zhang Z, Li ZH, Gao LB. et al. MicroRNA variants and colorectal cancer risk: a meta-analysis. Genetics and molecular research: GMR. 2016:15
72. Liu XX, Wang M, Xu D, Yang JH, Kang HF, Wang XJ. et al. Quantitative Assessment of the Association between Genetic Variants in MicroRNAs and Colorectal Cancer Risk. BioMed research international. 2015;2015:276410
73. Jia H, Yu H, Liu Q. Single nucleotide polymorphisms of MIR-149 gene rs2292832 contributes to the risk of hepatocellular carcinoma, but not overall cancer: a meta-analysis. Minerva medica. 2016;107:259-69
74. Wang XH, Wang FR, Tang YF, Zou HZ, Zhao YQ. Association of miR-149C>T and miR-499A>G polymorphisms with the risk of hepatocellular carcinoma in the Chinese population. Genetics and molecular research: GMR. 2014;13:5048-54
75. Feng Y, Duan F, Song C, Zhao X, Dai L, Cui S. Systematic evaluation of cancer risk associated with rs2292832 in miR149 and rs895819 in miR27a: a comprehensive and updated metaanalysis. Oncotarget. 2016;7:22368-84
76. Liu MF, Chen WQ, He YZ, Gu YL. Role of miR-149C>T polymorphisms on the risk of hepatocellular carcinoma in a Chinese population. Genetics and molecular research: GMR. 2014;13:7184-9
77. Sushma PS, Jamil K, Kumar PU, Satyanarayana U, Ramakrishna M, Triveni B. Genetic Variation in MicroRNAs and Risk of Oral Squamous Cell Carcinoma in South Indian Population. Asian Pac J Cancer Prev. 2015;16:7589-94
78. Wei WJ, Lu ZW, Li DS, Wang Y, Zhu YX, Wang ZY. et al. Association of the miR-149 Rs2292832 polymorphism with papillary thyroid cancer risk and clinicopathologic characteristics in a Chinese population. International journal of molecular sciences. 2014;15:20968-81
79. Wu J, Lv S, An J, Lu C. Pre-miR-149 rs71428439 polymorphism is associated with increased cancer risk and AKT1/cyclinD1 signaling in hepatocellular carcinoma. International journal of clinical and experimental medicine. 2015;8:13628-33
80. Tu HF, Liu CJ, Chang CL, Wang PW, Kao SY, Yang CC. et al. The association between genetic polymorphism and the processing efficiency of miR-149 affects the prognosis of patients with head and neck squamous cell carcinoma. PLoS One. 2012;7:e51606
81. Wang Z, Wei M, Ren Y, Liu H, Wang M, Shi K. et al. miR149 rs71428439 polymorphism and risk of clear cell renal cell carcinoma: a case-control study. Tumour Biol. 2014;35:12127-30
82. Li L, Liu T, Li Z, Zhang L, Zhang Z. The miR-149 rs2292832 T/C polymorphism may decrease digestive cancer susceptibility: an updated meta-analysis. International journal of clinical and experimental medicine. 2015;8:15351-61
83. Wang C, Sturgis EM, Chen X, Zheng H, Wei Q, Li G. Pre-miRNA variants as predictors of clinical outcome in patients with squamous cell carcinomas of the nonoropharynx. Oncotarget. 2016;7:26444-53
84. Huang GL, Lu Y, Pu XX, He YX, Chen ML, Li YZ. et al. Association study between miR-149 gene polymorphism and nasopharyngeal carcinoma. Biomedical reports. 2013;1:599-603
85. Lingzi X, Zhihua Y, Xuelian L, Yangwu R, Haibo Z, Yuxia Z. et al. Genetic variants in microRNAs predict non-small cell lung cancer prognosis in Chinese female population in a prospective cohort study. Oncotarget. 2016;7:83101-14
86. Hong MJ, Choi YY, Jang JA, Jung HJ, Lee SY, Lee WK. et al. Association between genetic variants in pre-microRNAs and survival of early-stage NSCLC. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2013;8:703-10
87. Wang F, Ma YL, Zhang P, Shen TY, Shi CZ, Yang YZ. et al. SP1 mediates the link between methylation of the tumour suppressor miR-149 and outcome in colorectal cancer. J Pathol. 2013;229:12-24
88. Safe S. MicroRNA-Specificity Protein (Sp) Transcription Factor Interactions and Significance in Carcinogenesis. Current pharmacology reports. 2015;1:73-8
89. Wilting SM, Verlaat W, Jaspers A, Makazaji NA, Agami R, Meijer CJ. et al. Methylation-mediated transcriptional repression of microRNAs during cervical carcinogenesis. Epigenetics. 2013;8:220-8
90. Oster B, Linnet L, Christensen LL, Thorsen K, Ongen H, Dermitzakis ET. et al. Non-CpG island promoter hypomethylation and miR-149 regulate the expression of SRPX2 in colorectal cancer. International journal of cancer. 2013;132:2303-15
91. Lin L, Zhang YD, Chen ZY, Chen Y, Ren CP. The clinicopathological significance of miR-149 and PARP-2 in hepatocellular carcinoma and their roles in chemo/radiotherapy. Tumour Biol. 2016;37:12339-46
92. Xue L, Wang Y, Yue S, Zhang J. Low MiR-149 expression is associated with unfavorable prognosis and enhanced Akt/mTOR signaling in glioma. International journal of clinical and experimental pathology. 2015;8:11178-84
93. Xu Y, Lin YP, Yang D, Zhang G, Zhou HF. Clinical Significance of miR-149 in the Survival of Patients with Laryngeal Squamous Cell Carcinoma. BioMed research international. 2016;2016:8561251
94. Li D, Chen P, Li XY, Zhang LY, Xiong W, Zhou M. et al. Grade-specific expression profiles of miRNAs/mRNAs and docking study in human grade I-III astrocytomas. Omics: a journal of integrative biology. 2011;15:673-82
95. Yang C, Sun C, Liang X, Xie S, Huang J, Li D. Integrative analysis of microRNA and mRNA expression profiles in non-small-cell lung cancer. Cancer gene therapy. 2016;23:90-7
96. Izquierdo L, Ingelmo-Torres M, Mallofre C, Lozano JJ, Verhasselt-Crinquette M, Leroy X. et al. Prognostic value of microRNA expression pattern in upper tract urothelial carcinoma. BJU international. 2014;113:813-21
97. Fogli S, Polini B, Carpi S, Pardini B, Naccarati A, Dubbini N. et al. Identification of plasma microRNAs as new potential biomarkers with high diagnostic power in human cutaneous melanoma. Tumour Biol. 2017;39:1010428317701646
98. Venkatesan N, Kanwar J, Deepa PR, Khetan V, Crowley TM, Raguraman R. et al. Clinico-Pathological Association of Delineated miRNAs in Uveal Melanoma with Monosomy 3/Disomy 3 Chromosomal Aberrations. PLoS One. 2016;11:e0146128
99. Kinoshita T, Yip KW, Spence T, Liu FF. MicroRNAs in extracellular vesicles: potential cancer biomarkers. J Hum Genet. 2017;62:67-74
100. van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: a common pathway for a specialized function. Journal of biochemistry. 2006;140:13-21
101. Tomasetti M, Lee W, Santarelli L, Neuzil J. Exosome-derived microRNAs in cancer metabolism: possible implications in cancer diagnostics and therapy. Experimental & molecular medicine. 2017;49:e285
102. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature cell biology. 2007;9:654-9
103. Tkach M, Thery C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell. 2016;164:1226-32
104. Pfeffer SR, Grossmann KF, Cassidy PB, Yang CH, Fan M, Kopelovich L. et al. Detection of Exosomal miRNAs in the Plasma of Melanoma Patients. Journal of clinical medicine. 2015;4:2012-27
105. Li J, Chen Y, Guo X, Zhou L, Jia Z, Peng Z. et al. GPC1 exosome and its regulatory miRNAs are specific markers for the detection and target therapy of colorectal cancer. Journal of cellular and molecular medicine. 2017;21:838-47
Author contact
![]() Corresponding author: Jinhai Tang, MD, PHD. Department of General Surgery, the First Affiliated Hospital with Nanjing Medical University, Nanjing 210029, P.R. China. E-mail address: tangjhedu.cn
Corresponding author: Jinhai Tang, MD, PHD. Department of General Surgery, the First Affiliated Hospital with Nanjing Medical University, Nanjing 210029, P.R. China. E-mail address: tangjhedu.cn

 Global reach, higher impact
Global reach, higher impact