3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(16):3343-3355. doi:10.7150/jca.20739 This issue Cite
Research Paper
The Prognostic Value of Peripheral Benzodiazepine Receptor in Patients with Esophageal Squamous Cell Carcinoma
1. Department of Anesthesiology, Cancer Center, Sun Yat-Sen University, State Key Laboratory of Oncology in South China, Guangzhou510060, Guangdong, China;
2. Department of Thoracic Oncology, Cancer Center, Sun Yat-Sen University, State Key Laboratory of Oncology in South China, Guangzhou510060, Guangdong, China;
3. Department of Hepatobiliary Oncology, Cancer Center, Sun Yat-Sen University, State Key Laboratory of Oncology in South China, Guangzhou510060, Guangdong, China.
* These authors contributed equally to this work.
Received 2017-4-25; Accepted 2017-8-6; Published 2017-9-20
Abstract
Background: The peripheral benzodiazepine receptor (PBR) has previously been reported as an oncogene in prostate, breast and colorectal cancers, but its prognostic value, biological behavior and function in esophageal squamous cell carcinoma (ESCC) has not been investigated.
Methods: qRT-PCR, western blotting and immunohistochemistry (IHC) were used to detect PBR expression in ESCC and matched non-cancerous tissues. Based on all of the significantly independent factors, a nomogram was established to predict the prognosis of ESCC patients. In addition, we performed comprehensive in vitro experiments to study the functions of PBR in cell growth, colony formation, and migration ability, as well as its relationship with epithelial-mesenchymal transition (EMT) related proteins in ESCC cells.
Results: The mRNA and protein expression levels of PBR in ESCC were higher than those in adjacent non-tumor esophageal epithelial tissues. The IHC results demonstrated that PBR expression was an independent prognostic factor in ESCC survival, patients with higher PBR expression had a poorer survival than those with low expression, and PBR expression was significantly associated with lymphoid nodal status. Furthermore, a nomogram was established to reliably predict the probability of death in ESCC patients, with a Harrell's c-index of 0.696. In the vitro experiments, knocking down the expression of PBR inhibited proliferation, colony formation and migration of ESCC cells, and regulated EMT-associated proteins (up-regulation of E-cadherin, ZO-1 and β-catenin and concomitant with down-regulation of Fibronectin and N-cadherin).
Conclusions: PBR is an independent prognostic factor in ESCC, and it promotes ESCC progression and metastasis. Basing on PBR expression level, a nomogram is established and performs a well in predicting survival of ESCC patients.
Keywords: Peripheral benzodiazepine receptor (PBR), Esophageal squamous cell carcinoma (ESCC), Prognosis, Nomogram.
Introduction
Esophageal cancer (EC) is the eighth most common cancer and the sixth leading cause of cancer-related deaths worldwide, with an estimated 455,800 new cases and 400,200 deaths in 2012, respectively [1, 2]. The incidence rate of esophageal cancer appears to be heterogeneous in terms of geographical areas and gender, with an occurrence of nearly 21:1 and 3-4:1, respectively [1]. There are two major histopathological types of EC: esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma. Factors contributing to EC include poor nutritional status, low intake of fruits and vegetables, drinking and smoking, and approximately 80% of all cases occur in less-developed regions, including Eastern Asia and Africa [1, 3]. In addition, as the predominant histopathological type in China, ESCC represents more than 90% of new cases, and its incidence and mortality has reached to half of the world [3, 4]. Despite the improvement in the surgical approaches and the multidisciplinary treatments, the 5-year survival rate remains poor [5]. Therefore, there is an urgent clinical need to develop new molecular targets to predict the prognosis and guide therapeutic strategies.
The Translocator protein (TSPO) gene, located on the 22q13.31 chromosome, encodes a widely expressed 18 kDa mitochondrial protein, peripheral benzodiazepine receptor (PBR) [6-8]. PBR was first identified in 1977, as an alternative binding site in kidney for the benzodiazepine diazepam [9]. More and more new functions of PBR were studied and elaborated on over the following decades. PBR is a translocator that imports cholesterol into mitochondria, which is related to its important function in steroidogenesis [10]. In addition, PBR can directly or indirectly regulate multiple cellular functions, including apoptosis, cell proliferation, differentiation, anion transport, porphyrin transport, heme synthesis, immune response and regulation of mitochondrial function [7, 11-13]. Based on these functions, PBR expression has been associated with various diseases, including brain injury, neurodegeneration, ischemia-reperfusion and cancer [14]. In terms of the relationship between tumors and PBR, there is clear evidence showing that the expression level of PBR is elevated in brain gliomas, breast cancer, colon adenocarcinoma, hepatocellular carcinoma, oesophageal carcinoma, prostate cancer, and endometrial and ovarian carcinomas, compared to the normal tissues [15-18], which may also indicated that PBR has a role in carcinogenesis. Important steps in tumorigenesis include infinite proliferative capability and loss of apoptotic ability. Indeed, PBR ligands can inhibit the proliferation of cancerous breast, melanoma, testis, colon, prostate and astrocytoma cells [19-24]. At the same time, PBR ligands have been shown to induce apoptosis in melanoma, hepatocellular, breast, oesophageal and colon carcinoma cell lines [21, 25-27]. However, until now, both the proliferative and antiapoptotic role of PBR in solid tumor cells have only been verified using PBR drug ligands, rather than through direct evidence [25, 26, 28]. Only in astrocytomas, prostate, breast and colorectal cancers, there is direct evidence to indicate that PBR expression is a prognostic factor and a potential therapeutic target [15, 29-31].
In past years, only using PBR ligands, the importance of PBR in regulating the apoptosis of esophageal carcinoma cell lines has been preliminarily reported [26, 28]. However, no evidence has been provided regarding the prognostic value of PBR and no direct evidences has been observed regarding the cellular functions and molecular mechanism of PBR in ESCC. Hence, we decided to examine the differential expression of PBR at the mRNA and protein levels, in adjacent non-cancerous and ESCC tissues to evaluate its prognostic value using immunohistochemistry, establish a simple and easily visualized clinical nomogram, and verify the function of PBR in ESCC cell lines. Intriguingly, in addition to its prognostic significance, we also found that knockdown of PBR could inhibit the proliferation, colony formation ability, migration ability, and epithelial-mesenchymal transition (EMT) related proteins in ESCC cell lines.
Patients and Methods
Patients and tissue samples
All tissues were collected from ESCC patients who underwent radical esophagectomy at Sun Yat-sen University Cancer Center between October 2000 and April 2007, and tissue samples included 205 esophageal squamous cell carcinoma (ESCC) samples and 141 paired adjacent non-cancerous esophageal tissues. An additional 28 ESCC and paired adjacent non-tumor esophageal tissues, examined by quantitative RT-PCR and western blotting, were stored in liquid nitrogen during 2016. Those tissues also came from patients, who were pathologically and clinically diagnosed ESCC and were treated with radical esophagectomy without neoadjuvant and/or adjuvant chemoradiotherapy. All patients in the study had complete medical and follow-up records. The tumor pathological stage was based on the 7th edition of the Union for International Cancer Control-TNM Classification. This study was approved by the medical committee of the Sun Yat-sen University Cancer Centre (No.YB2016-056).
Human esophageal squamous cell lines
The ESCC cell lines KYSE140 and KYSE410 were kindly provided by Professor Mu-Sheng Zeng (State Key Laboratory of Oncology in South China, Sun Yat-sen University Cancer Center) [32]. All ESCC cell lines were cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT) and in humidified conditions with 5% CO2 and at 37°C.
RNA isolation and Quantitative RT-PCR
Total RNA was isolated from ESCC tissue samples and cell lines using TRIzol reagent (Invitrogen), according to the manufacturer's instructions. The quality and concentration of the RAN were examined, with a NanoDrop spectrophotometer (ND-1000, Thermo Scientific, Massachusetts, USA). Complementary DNA (cDNA) was synthesized using 2 µg of RNA, according to the instructions provided in the reverse transcriptase kit (Invitrogen). Quantitative RT-PCR was performed using SYBR Green SuperMix on an ABIPrism-7500 Sequence Detector System (ABI, Applied Biosystems, Carlsbad, USA) to measure the mRNA level of the target genes. The expression data were normalized to the housekeeping gene (β-actin), which was used as an internal control. After normalization, we used 2-ΔΔCt normalization to calculate the mRNA copy number ratios. The primer sequences were as follows: PBR sense: 5'- CGGCCTGGCTAACTCCTG-3', PBR antisense: 5'-CTGCAGCTGCTGTTCAGGG-3', β-actin sense: 5'- CACCATTGGCAATGAGCGGTTC-3', β-actin antisense: 5'- AGGTCTTTGCGGATGTCCACGT-3'. To ensure the repeatability and accuracy of the results, all genes were tested three times.
Western blotting analysis
Equal amounts of tissues or cell lysates (30µg per lane) were electrophoretically separated by 10% SDS polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membrane (Pall, Port Washington, USA). The membrane was blocked with 5% skimmed milk for 1 hour at room temperature, incubated with the indicated antibodies [α-tublin (1:3000, Santa Cruz Biotechnology, USA), PBR (1:1000, Abcam, USA), E-cadherin (1:1000, BD Biosciences, USA), N-cadherin (1:1000, BD Biosciences, USA), Fibronectin (1:1000, BD Biosciences, USA), β-catenin (1:1000, BD Biosciences, USA), ZO-1(1:1000, Invitrogen, USA)] overnight at 4°C, and incubated for 45 min with the appropriated anti-rabbit or anti-mouse antibody.
Immunohistochemical (IHC) analysis
All enrolled paraffin-embedded specimens were cut into 4 µm thick sections. IHC staining was performed using the standard method of the Dako Envision system. Briefly, all sections were deparaffinized, rehydrated and blocked of endogenous peroxidase activity using 0.3% hydrogen peroxide for 15 minutes. Then, the sections were microwave-treated in 10 mM citrate buffer (pH 6.0) for 10 minutes. After natural cooling, the sections were incubated with anti-PBR monoclonal antibody (1:6000, Abcam, USA) at 4°C overnight and with a secondary antibody at 37°C for 30 minutes on the next day. Subsequently, diaminobenzidine was used to stain the target protein, and Mayer's hematoxylin was applied to redye the cell nuclei. All stained sections were assessed and scored independently by two senior pathologists (Dr. Mei Li and Dr. Rong-Zhen Luo), who were blinded to the clinical parameters. The final immunoreactivity score (IRS) was calculated by multiplying of the staining intensity about staining (0: no, 1: weak, 2: moderate or 3: strong) and the percentage of positively stained cells (0: 0-5%; 1: 6%-25%; 2: 26%-50%; 3: 51%-75%; 4:76%-100%) [33]. In our study, the median value of all IRS scores (8.0) was chosen as the optimistic cut-off point to determine the expression level of PBR in ESCC.
SiRNA transfection
The two siRNAs targeting the PBR [GenBank: NM_000714.5] were represented as siPBR#1 (5'- UGGGAGGCUUCACAGAGAA-3') and siPBR#2 (5'- CCUUCACGACCACACUCAA -3'). siNC, which was nonhomologous to any human genome sequences, was used as the negative control. The KYSE140 and KYSE410 cells were transfected with 50 nM RNA duplex and 5 µl of Lipofectamine RNAiMAX (Invitrogen), according to the manufacturer's instructions. All cells were cultured for 48 hours after transfection and then harvested for further experiments.
Cell growth assay in vitro
After transfection using siNC or siPBR#1 or siPBR#2 for 48 hours, KYSE140 (1000 cells/well) or KYSE410 (1200 cells/well) were seeded in 96-well plates in quintuplicate, and the cell growth rate was assayed with CCK-8 kit (Dojindo, Japan), according to the manufacturer's instructions. Subsequently, the absorbance of the solution after 2 hours was read at 450 nm using a Spectramax M5 microplate reader (Molecular Devices, Sunnyvale, USA).
Colony formation assay
Cells (1000 cells/well) were evenly seeded into 6-well plates. After being cultured for 14 days, the colonies were fixed with methanol for 10 minutes, stained with 0.5% crystal violet in 20% methanol and counted. To ensure accuracy, each group of cells was texted in triplicate.
Transwell assay
KYSE140 or KYSE410 (15×104 and 10×105 cells, respectively) cells in 200 µl of FBS-free RPMI were seeded in the top of a Transwell (BD Biosciences, San Jose, USA) chamber 48 hours post-transfection, while the lower chambers were filled with DMEM with 10% FBS. After 24 hours of incubation, cells on the lower surface of the chamber were fixed, stained and counted. Independent experiments were performed in triplicate.
Statistical analysis
All statistical analysis was performed using the SPSS (version 20.0) statistical software package (SPSS Inc., Chicago, IL, USA). Student's tests were used to estimate the significance of differences. Kaplan-Meier plots and log-rank tests were performed to analyze patient survival. The Cox proportional hazards model was implemented with univariable and multivariable analysis. The Pearson χ2 test was used to analyze the relationship between PBR expression and the clinicopathological characteristics. Two-tailed Mann-Whitney tests were performed to assess the differences between groups. A nomogram for possible prognostic factors associated with survival was established by R software, and the Harrell's concordance index (c-index) was used to evaluate the predictive accuracy (Supplemental material). p < 0.05 was considered to be statistically significant.
Results
Baseline characteristics of patients
The baseline characteristics of 205 ESCC patients, categorized by PBR expression are listed in Table 1.
The up-regulated expression of PBR in ESCC tissues
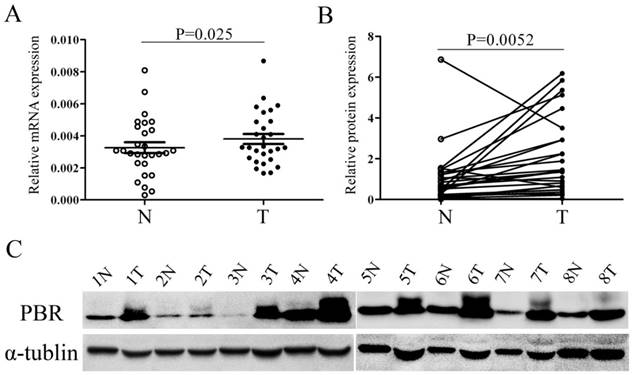
Quantitative real-time PCR (qRT-PCR) and western blotting were used to compare the PBR expression in ESCC tissues and corresponding non-cancerous esophageal mucosa derived from 28 ESCC patients. Both the protein expression of PBR and the PBR mRNA level, the expression of PBR in ESCC tissues was significantly higher than those in adjacent normal tissues (p=0.025 and p=0.0052, respectively, Fig. 1). The results of further immunohistochemistry staining in 141 matched ESCC and normal tissues were consistent with the differential expression observed in qRT-PCR and western-blot, with p <0.001. (Fig.2)
The association between PBR expression and progression of ESCC
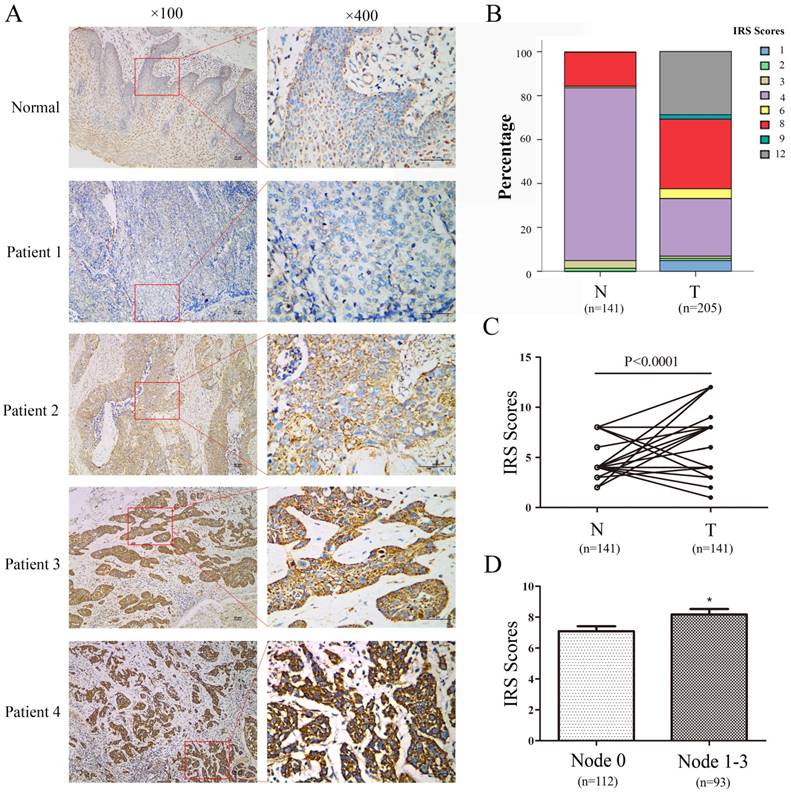
Sequentially, IHC staining was performed on 205 ESCC and 141 corresponding adjacent non-tumor esophageal tissues with a monoclonal PBR antibody. First, PBR staining was primarily displayed in the cytoplasm (Fig.2). Second, according to the IHC score criteria, as described above, the median IRS score in ESCC tissues was 8, which was typically higher than the score in matched normal tissues (median: 4, Fig.2). Though non-negative staining was detected in the normal group, most normal esophageal squamous epithelium (84.4%, 119/141) showed weak staining. In addition, most ESCC tissues were strongly stained, with a 62.4% (128/205) optimistic staining rate. The difference between IRS scores of ESCC and those matched normal tissues was statistically significant, and the staining in ESCC tissues was stronger than those in normal tissues. (p<0.001, Fig. 2)
Association between PBR expression and clinicopathological variables in 205 ESCC patients
| Variables | Cases | PBR expression | ||
|---|---|---|---|---|
| Low | High | P valuea | ||
| Age(years) | 0.109 | |||
| Medianb | 57 | |||
| Range | 32-80 | |||
| ≤57 | 105 | 45(42.9) | 60(57.1) | |
| >57 | 100 | 32(32.0) | 68(68.0) | |
| Gender | 0.199 | |||
| Male | 149 | 52(34.9) | 97(65.1) | |
| Female | 56 | 25(44.6) | 31(55.4) | |
| Smoking Status | 0.464 | |||
| Non-smoker | 76 | 31(40.8) | 45(59.2) | |
| Smoker | 129 | 46(35.7) | 83(64.3) | |
| Alcohol Intake | 0.550 | |||
| No | 155 | 60(38.7) | 95(61.3) | |
| Yes | 50 | 17(34.0) | 33(66.0) | |
| Tumor location | 0.934 | |||
| Upper | 15 | 5(33.3) | 10(66.7) | |
| Middle | 94 | 36(38.3) | 58(61.7) | |
| Lower | 96 | 36(37.5) | 60(62.5) | |
| Differentiation | 0.690 | |||
| G1 | 54 | 27(50.0) | 27(50.0) | |
| G2 | 105 | 32(30.5) | 73(69.5) | |
| G3 | 46 | 18(39.1) | 28(60.9) | |
| pT status | 0.628 | |||
| pT1 | 5 | 1(20.0) | 4(80.0) | |
| pT2 | 59 | 24(40.7) | 35(59.3) | |
| pT3 | 141 | 52(36.9) | 89(63.1) | |
| pN status | 0.045 | |||
| pN0 | 112 | 49(43.8) | 63(56.3) | |
| pN1-3 | 93 | 28(30.1) | 65(69.9) | |
| pTNM status | 0.051 | |||
| II | 129 | 55(42.6) | 74(57.4) | |
| III | 76 | 22(28.9) | 54(71.1) | |
aChi-square test; bmedian age was 57 years for 205 enrolled ESCC patients.
G, grade; pT, pathologic tumor; pN, pathologic node; pTNM, pathologic tumor-node-metastasis.
Combined with the further analysis of the immunohistochemistry results, another interesting observation was noted regarding the association between PBR expression and lymphoid nodal metastasis of ESCC patients. ESCC patients with lymphoid nodal metastasis had a higher expression level of PBR than those without lymphoid nodal metastasis (p= 0.045, Table 1). Regrettably, the p value, based on Pearson χ2 analysis of the PBR expression and TNM stage, was 0.051, and there were no significant associations between the PBR expression level and other clinicopathological variables (age, gender, smoking status, alcohol intake, tumor location, differentiation, pT status) (Table 1). These results indicated that PBR may play an important role in ESCC development and progression.
The correlation between PBR expression and ESCC survival
Further analysis of the complete clinicopathological and follow-up data was performed, including the median observation period, disease-free survival (DFS) and overall survival (OS), which were 57 months (range: 3-168 months), 43 months and 56 months, respectively. At the final clinical follow-up point, a total of 126 cancer-related deaths had occurred, and the 1-year, 3-year, and 5-year survival rate were 83.4%, 56.6% and 49.3%, respectively.
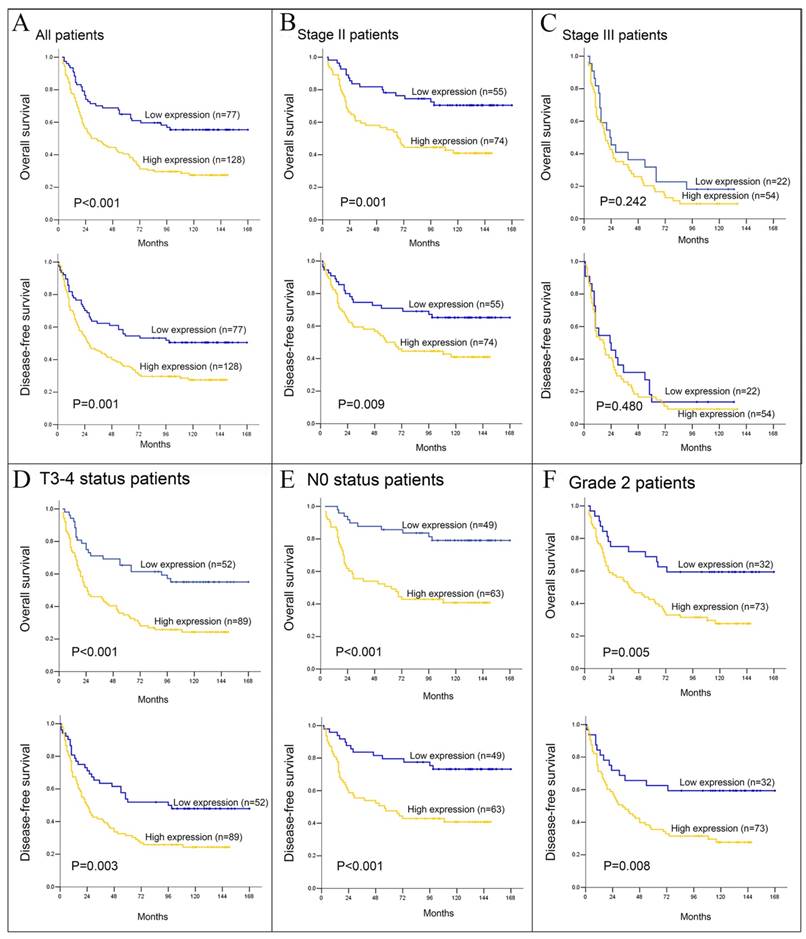
For the whole cohort, the values of survival times (both DFS and OS) among PBR-negative patients were higher than those among PBR-positive patients (median 54 vs. 18 months and 70 vs. 21 months, p=0.001 and p<0.001, respectively, Fig. 3). In the further stratification of patients groups based on tumor stage, in addition to the OS in tumor Grade 1 stratified by PBR expression (p=0.015), the PBR expression also showed further differences in DFS and OS when stratified by pT3-4 (p=0.003 and p<0.001, respectively), pN0 (both P<0.001), pTNM II stage (p=0.009 and p=0.001, respectively) and tumor grade 2 (p=0.008 and p=0.005, respectively, Table 2). But no significant association between PBR expression and survival was detected in other variables.
Furthermore, we established and analyzed a Cox proportional hazards model to further validate that whether PBR expression is an independent prognostic factor in ESCC. We entered a series of factors, including age, gender, smoking status, alcohol intake, tumor location, surgery methods, differentiation, pTNM stage and PBR expression, into the univariate Cox regression analysis. Only alcohol intake, differentiation, pTNM stage and PBR expression were associated with DFS and OS in a univariate analysis. Interestingly, the multivariate analysis model demonstrated that differentiation, pTNM stage and PBR expression were independent predictors in ESCC after enrolling all statistical variables from the univariate analysis (alcohol intake, differentiation, pTNM stage and PBR expression) (Table 3).
Nomogram development based on PBR expression
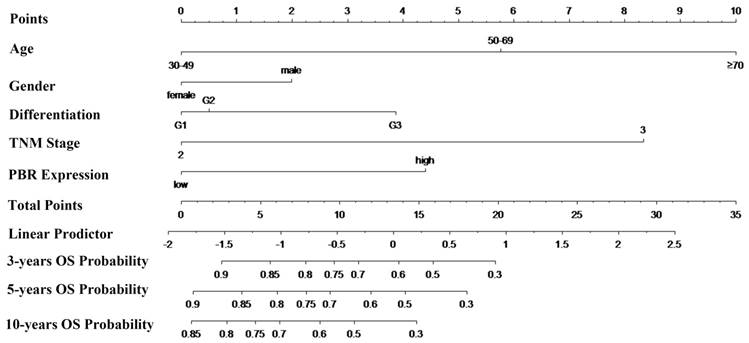
To predict the risk for patients with ESCC after radical esophagectomy, a novel nomogram model was established using prognostic factors (PBR expression, pTNM stage and differentiation) combined with age and sex (Fig. 4). Specifically, total points were identified to determine the survival probability for ESCC patients after esophagectomy, which were counted by summing of the top scale corresponding to each factor. In addition, the Harrell's c-index of the nomogram for OS prediction was 0.696.
PBR expression is up-regulated in esophageal squamous cell carcinoma tissues, compared with peritumoral normal esophageal tissues. The relative expression of PBR mRNA (A) and protein (B, C) was determined by quantitative RT-PCR and western blotting, respectively, in 28 pairs of matched ESCC and non-tumor tissues. mRNA levels are presented as the means± SD and normalized to the housekeeping gene β-actin in qRT-PCR. N, matched noncancerous tissue; T, tumor tissues; ESCC, esophageal squamous cell carcinoma.
PBR expressions in ESCC and peritumoral normal esophageal tissues were examined by immunohistochemistry. (A) Representative immunohistochemical results of PBR expression in normal esophageal epithelium tissue (weak staining) and four ESCC tissues (patient 1: negative staining, patient 2: weak staining, patient 3: moderate staining, patient 4: strong staining). (B) The relative percentage of staining was evaluated in paired ESCC tissues using immunoreactivity scores (IRS). (C) Significant difference was found between 114 paired ESCC tissues. (D) PBR expression in cases with lymphoid nodal metastasis was significantly higher than in cases without lymphoid nodal metastasis.
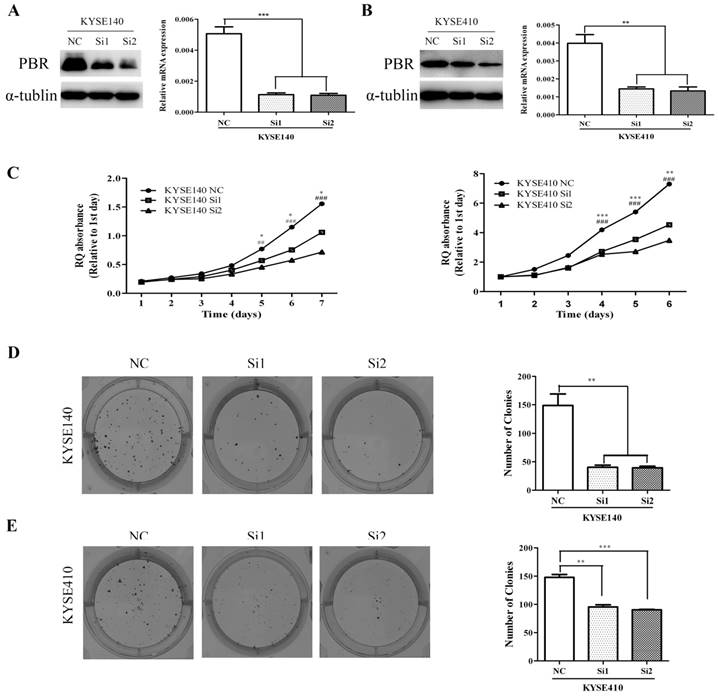
Down-regulation of PBR expression suppresses the proliferation and clonogenicity of ESCC cell lines
To investigate the functional role of PBR in ESCC tumorigenesis, we knocked down the PBR expression in KYSE140 and KYSE410 cells using two siRNA duplexes. In addition, the PBR expression was confirmed to be down-regulated by qRT-PCR and Western blotting assay (Fig. 5). Later, we used an MTT assay to detect the proliferative ability of ESCC cells. Compared to the negative control (NC), siPBR treatments could markedly reduce the proliferation rate of ESCC cells (Fig. 5). Moreover, down-regulated PBR expression in esophageal squamous cells KYSE140 and KYSE410 caused a dramatic decrease in both the size and number of colonies (Fig. 5). Together, our findings suggested that PBR could play a growth-promoting role in ESCC cells.
Kaplan-Meier survival analysis, in related to PBR expression, was performed in ESCC patients. Overall survival (OS) and disease-free survival (DFS) for all 205 ESCC (A), stage II (B) and III (C) patients was further analyzed. (D, E and F) OS and DFS for T3-4, N0 and Grade 2 ESCC patients, respectively.
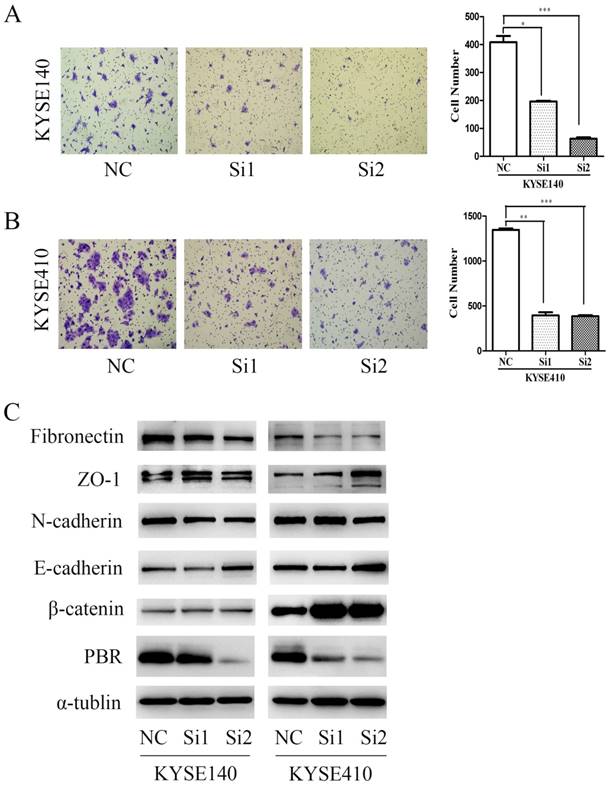
Knockdown of PBR expression inhibited migration of ESCC cells
To confirm the correlation between PBR and metastasis of ESCC, we performed and compared Transwell assays with KYSE140 and KYSE410 cells. After a 24-hour incubation, the number of cells that migrated in the groups with siPBR treatments was significantly less than in the NC group (Fig. 5), which is consistent with the previous clinical results. As is well known, EMT is a trigger that activates the invasion ability of cancer cells, and its physiological relationship with carcinogenesis and cancer metastasis has been verified by an accumulation of observations in human tumors and experimental animal models [34-36]. To study the possible mechanism of PBR in terms of ESCC migration, we examined and compared the expression level of EMT-associated proteins in KYSE140 and KYSE410 cells with siPBR treatment and the NC group. Intriguingly, we found that knocking down the PBR expression in ESCC cells can up-regulate the expression of E-cadherin, ZO-1 and β-catenin, and concomitantly down-regulated the expression of Fibronectin and N-cadherin expression (Fig. 6). These results indicated that the mechanism of PBR may involve EMT regulation in ESCC cells.
Discussion
In spite of the abundance of studies on PBR reported in the past several decades, the functions of PRB are not yet fully understood. Recently, several studies have reported that PBR is ubiquitously expressed in most healthy and diseased tissues, but its expression varies considerably. For example, PBR is expressed at a high level in steroid-producing, secretory and glandular tissues [37]. Meanwhile, intermediate levels of PBR are detected in renal and myocardial tissues, and low levels are found in brain, liver, skeletal muscle and the gastrointestinal tract [7, 37]. These findings only focused on the protein levels of PBR in some tissues, but they also indicated that heterogeneity of PBR expression may be related to different cellular functions. Therefore, in the last decade, more and more studies have fueled an interest in the function of PBR in relation to cancer, making it necessary to discuss recent advances about complex functions of PBR.
Prognostic value of PBR expression in 205 ESCCs (log- rank test)
| PBR expression | Cases | DFS(months) | OS(months) | ||||
|---|---|---|---|---|---|---|---|
| Mean | Median | P-valuea | Mean | Median | P-valuea | ||
| Total | 205 | 0.001 | <0.001 | ||||
| Low expression | 77 | 98 | NR | 108 | NR | ||
| High expression | 128 | 60 | 27 | 62 | 29 | ||
| pT status | |||||||
| pT1-2 | 64 | 0.161 | 0.140 | ||||
| Low expression High expression | 25 39 | 99 72 | NR 56 | 103 75 | NR 66 | ||
| pT3-4 | 141 | 0.003 | <0.001 | ||||
| Low expression | 52 | 96 | 96 | 108 | NR | ||
| High expression | 89 | 54 | 24 | 57 | 25 | ||
| pN status | |||||||
| pN0 | 112 | <0.001 | <0.001 | ||||
| Low expression | 49 | 134 | NR | 142 | NR | ||
| High expression | 63 | 77 | 56 | 78 | 63 | ||
| PN1-3 | 93 | 0.414 | 0.918 | ||||
| Low expression | 28 | 33 | 13 | 44 | 23 | ||
| High expression | 65 | 41 | 22 | 46 | 25 | ||
| pTNM | |||||||
| Stage II | 129 | 0.009 | 0.001 | ||||
| Low expression | 55 | 120 | NR | 130 | NR | ||
| High expression | 74 | 80 | 58 | 82 | 67 | ||
| Stage III | 76 | 0.480 | 0.242 | ||||
| Low expression | 22 | 38 | 23 | 47 | 23 | ||
| High expression | 54 | 31 | 17 | 36 | 19 | ||
| Differentiation | |||||||
| G1 | 54 | 0.067 | 0.015 | ||||
| Low expression | 27 | 112 | NR | 123 | NR | ||
| High expression | 27 | 70 | 66 | 71 | 66 | ||
| G2 | 105 | 0.008 | 0.005 | ||||
| Low expression | 32 | 109 | NR | 113 | NR | ||
| High expression | 73 | 61 | 33 | 64 | 40 | ||
| G3 | 46 | 0.470 | 0.165 | ||||
| Low expression | 18 | 46 | 26 | 59 | 28 | ||
| High expression | 28 | 41 | 15 | 44 | 21 | ||
aLog- rank test.
DFS, disease-free survival; OS, overall survival; pT, pathologic tumor; pN, pathologic node; pTNM, pathologic tumor-node-metastasis; G, tumor grade; NR, not reached.
Univariate and multivariate regression analysis for DFS and OS in the whole cohort
| Disease-free survival | Overall survival | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | |||||||||
| HR | 95% CI | P valuea | HR | 95% CI | P valuea | HR | 95% CI | P valuea | HR | 95% CI | P valuea | |
| Ageb | 1.212 | 0.858-1.710 | 0.277 | … | … | … | 1.275 | 0.898-1.809 | 0.175 | … | … | … |
| Genderc | 0.767 | 0.517-1.139 | 0.189 | … | … | … | 0.706 | 0.470-1.059 | 0.092 | … | … | … |
| Smoked | 1.208 | 0.843-1.731 | 0.303 | … | … | … | 1.246 | 0.863-1.797 | 0.240 | … | … | … |
| Alcohol Intakee | 1.517 | 1.039-2.215 | 0.031 | 1.380 | 0.942-2.020 | 0.098 | 1.584 | 1.082-2.319 | 0.018 | 1.453 | 0.990-2.134 | 0.057 |
| Locationf | 0.969 | 0.734-1.280 | 0.824 | … | … | … | 0.974 | 0.734-1.293 | 0.855 | … | … | … |
| Surgeryg | 1.026 | 0.857-1.229 | 0.780 | 1.450 | … | … | 1.018 | 0.848-1.222 | 0.846 | … | … | … |
| Differentiationh | 1.545 | 1.204-1.982 | 0.001 | 2.792 | 1.112-1.890 | 0.006 | 1.511 | 1.175-1.942 | 0.001 | 1.386 | 1.059-1.814 | 0.018 |
| TNM stage i | 3.323 | 2.334-4.730 | <0.001 | 1.558 | 1.949-3.999 | <0.001 | 3.488 | 2.439-4.988 | <0.001 | 2.863 | 1.988-4.122 | <0.001 |
| PBRj | 1.823 | 1.248-2.663 | 0.002 | 1.063-2.284 | 0.023 | 2.161 | 1.456-3.207 | <0.001 | 1.879 | 1.294-2.730 | 0.001 | |
aCox proportional hazards model; bAge≤57 vs. Age>57; cMale vs. Female; dSmoker vs. Non-smoker; eDrinkers vs. Non-drinkers; fUpper thoracic vs. Middle thoracic vs. Lower thoracic; gLeft thoracotomy vs. Thoracic-abdominal-cervical incision; h Tumor grade 1 vs. Tumor grade 2 vs. Tumor grade 3; ipTNM stage II vs. pTNM stage II; jHigh expression of PBR vs. low expression of PBR.HR, hazard ratio; CI, confidence interval.
Nomogram model for the probability of three-, five- and ten- years overall survival (OS) predictions. The nomogram is used by determining the total points identified by summation of the corresponding points corresponding to each of the factors. The Harrell's c-index for OS prediction was 0.696.
Knockdown of PBR suppresses the proliferation and clone formation of ESCC cells. PBR expression in KYSE140 (A) and KYSE410 (B) cells transfected with NC or PBR-targeting siRNAs was down-regulated α-tublin was used as an internal control. (C) Growth curves of KYSE140 and KYSE410 cells transfected with NC or PBR-targeting siRNAs. Colony formation assay of KYSE140 (E) and KYSE410 (F) cells infected with NC or PBR-targeting siRNAs. (*/#p<0.05, **/##p<0.01, ***/###p<0.001)
PBR silencing inhibited ESCC cell migration. Transwell assay of KYSE140 (A) and KYSE410 (B) cells transfected with NC or PBR-targeting siRNAs. Data represent the means± SD. Data presented as the means± SD. (C) Western blots of ESCC cell lines (KYSE140 and KYSE410) with NC and PBR-targeting siRNA were analyzed, with α-tublin as an internal control. (*p<0.05, **p<0.01, ***p<0.001)
From the history of PBR studies in cancer, the previous studies have determined that PBR expression is elevated in several cancerous tissues, including brain gliomas, breast cancer, colon adenocarcinoma, hepatocellular carcinoma, esophageal carcinoma, prostate cancer, and endometrial and ovarian carcinomas, suggesting a potential role of PBR in carcinogenesis [16, 18]. Further studies found that the proliferative and anti-apoptotic capacity of PBR is indirectly verified by PBR ligands in some cancers [25, 26, 28]. To date, there has been some direct evidence to confirm that PBR could play a role as a negative predictor of survival in astrocytomas, prostate, breast and colorectal cancers [15, 29-31].
Recently, studies have reported that the differential gene expression of PBR in 17 pairs ESCC and normal tissues and PBR ligands induce apoptosis in ESCC cells [26, 28], but the prognostic value of PBR in ESCC has not been investigated. First, we confirmed the differences between mRNA and protein expression of PBR between in a relatively large scale of tumor and normal tissues, and our results are in accordance with previous finding. Second, the result of immunohistochemistry staining reconfirmed the differential expression observed in qRT-PCR and western-blot. Furthermore, we found that the immunohistochemical staining of PBR in ESCC displayed a cytoplasmic pattern, rather than nuclear nor membranous pattern, this staining pattern is in agreement with the results in astrocytomas and colorectal cancer [15, 31], but is different from the staining in breast and prostate carcinoma, which were observed in both the cytoplasm and nucleus [30]. Some research also found that nuclear PBR is responsible for regulating cholesterol movement and proliferation of cancer cells [30, 38]. Together, those results indicated that the PBR staining pattern could be related to its different functions in different cancers.
Combined with the further statistical analysis, we found that PBR expression was positively associated with the number of lymphoid nodal metastases in ESCC, which indicated a correlation between PBR and progression of ESCC. This is the first time a relationship between PBR and lymphoid nodal metastasis of cancer has been found. Zeqiu et al. found that high-grade human breast cancer had higher PBR expression than low-grade cancer [30]. However, we failed to find any other correlation between PBR expression and other clinicopathological variables, including differentiation, age, gender and pTNM stage. A potential reason for this phenomenon is the heterogeneity of biomarkers in different types of tissues. Furthermore, one of the most important findings is that PBR plays a role as an independent prognostic factor in ESCC, and patients with higher PBR expression had a poorer survival sate than those with low-expression, especially in the pTNM II stage. These results indicated that in addition to the prognostic value of PBR, it could also be useful as a biomarker for earlier diagnosis of ESCC.
It is well known that a nomogram can establish a simple and visual graphic representation of a statistical predictive model. More importantly, the accuracy and usability of nomograms are accepted by more and more clinics. Therefore, we attempted to establish a nomogram to predict the probability of death in ESCC patients based on PBR expression, differentiation, and pTNM stage combined with age and sex. More importantly, this nomogram performed well in predicting about OS, because its c-index is 0.696, which is the second promising finding.
To further investigate the biological significance and functions of PBR in ESCC cell lines, we knocked down PBR expression in two ESCC cell lines using two siRNA duplexes. We found that ESCC cells had a dramatically lower proliferation rate, a smaller number of colonies, and less migration ability after siPBR treatments. Together, these results gave us more reasons to believe that PBR might be an oncogene in ESCC. Reviewing the previous results regarding the association between PBR and lymphoid nodal metastasis, suggests that the function of PBR has a large effect on the metastasis of ESCC. At the same time, epithelial-mesenchymal transition (EMT) is a trigger that leads to dissociation of carcinoma cells from primary carcinomas, followed by migration and dissemination to distant sites.[34] Therefore, we thought that PBR may influence the metastasis by regulating EMT-related proteins in ESCC. To examine and confirm this hypothesis, we compared the expression of EMT-associated proteins in the negative control and siPBR treatment ESCC cells. Intriguingly, knocking down PBR expression in ESCC cell lines can caused up-regulation of Z0-1, β-catenin and E-cadherin, accompanied by the down-regulation of Fibronectin and N-cadherin. Although other previous papers noted PBR function in cancer metastasis and examined the expression of PBR and Vimentin, they failed to note the relationship between PBR and EMT other related proteins, perhaps because of the experimental limitations of using only one EMT-associated protein (Vimentin) [11]. PBR regulates EMT-associated proteins in ESCC, which could be a potential mechanism underlying its biological behaviors and prognostic value, and a potential trigger to promote lymphoid nodal metastasis. Despite the fact that PBR has been known for several decades, the function of PRB in cancers is not yet fully understood. Previous studies have almost always focused on the relationship between PBR and apoptosis [25, 26, 28]. The third remarkable point is that this is the first reported finding that EMT-associated proteins are regulated by PBR in ESCC, which maybe a potential mechanism to explain PBR as a predictor for ESCC patients. Of course, the exact mechanisms require further experimental exploration.
Some previous reports showed that the expression of PBR could influence chemosensitivity in some cancers [39], but we failed to clarify whether this relationship exists in ESCC, because our study was limited to enrolled patients who underwent radical esophagectomy without chemotherapy. However, some previous studies have suggested the importance of EMT in conferring chemoresistance in breast and pancreatic cancer models [40, 41], which seems to provide a theoretical possibility for our detailed investigation. Our study has two other potential limitations. First, as a retrospective study, the number of patients enrolled in our study was not very large. Second, the c-index of the nomogram shows that the model has a good, but not perfect predictive ability, and it would be better to use an external study cohort to validate this nomogram. Therefore, further studies are needed to confirm and perfect these preliminary results.
In conclusion, we firstly confirmed that PBR is more highly expressed in ESCC than in adjacent non-cancerous esophageal squamous epithelium at mRNA and protein levels, and we found that ESCC patients with higher PBR levers exhibited a substantially poorer survival rate. In addition, we firstly found that PBR is an independent prognostic factor in ESCC using histochemistry analysis. Moreover, by enrolling PBR expression and any other the independent prognostic factors, a nomogram was firstly established to predict the probability of death for ESCC patients, with a good predictive ability. Besides that, knocking down PBR contributed to decreased proliferation, colony formation, and migration in ESCC cells, and also regulated EMT-associated proteins. Taken together, our study provides convincing evidences to support PBR promotes ESCC progression and metastasis using clinical data and vitro experiments. Furthermore, this is the first time a nomogram based on PBR expression level, which performs a well in predicting survival and could directly be used to guide clinical treatment, has been established.
Abbreviations
PBR, peripheral benzodiazepine receptor; TSPO, translocator protein; EC, esophageal cancer; ESCC, esophageal squamous cell carcinoma; EMT, epithelial-mesenchymal transition; IHC, immunohistochemistry; IRS, immunoreactivity score; qRT-PCR, quantitative real-time PCR; DFS, disease-free survival; OS, overall survival.
Supplementary Material
Related computerized programs for the nomogram using R software in our study.
Acknowledgements
All authors are very grateful to Prof. Mu-Sheng Zeng for providing the ESCC cell lines (KYSE140 and KYSE410 cell lines) and two pathologises (Dr. Mei Li and Dr. Rong-Zhen Luo) for assessing the staining of immunohistochemistry. This study was supported by the National Natural Science Foundation of China (No. 81271246 and No. 81372568), Guangdong Esophageal Cancer Institute (No.Q201606), and the State Scholarship Foundation of the China Scholarship Council (No. 201606380100).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108
2. van Hagen P, Hulshof MC, van Lanschot JJ, Steyerberg EW, van Berge Henegouwen MI, Wijnhoven BP. et al. Preoperative chemoradiotherapy for esophageal or junctional cancer. N Engl J Med. 2012;366:2074-84
3. Arnold M, Soerjomataram I, Ferlay J, Forman D. Global incidence of oesophageal cancer by histological subtype in 2012. Gut. 2015;64:381-7
4. Zhao J, He YT, Zheng RS, Zhang SW, Chen WQ. Analysis of esophageal cancer time trends in China, 1989- 2008. Asian Pac J Cancer Prev. 2012;13:4613-7
5. Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241-52
6. Anholt RR, Pedersen PL, De Souza EB, Snyder SH. The peripheral-type benzodiazepine receptor. Localization to the mitochondrial outer membrane. J Biol Chem. 1986;261:576-83
7. Batarseh A, Papadopoulos V. Regulation of translocator protein 18 kDa (TSPO) expression in health and disease states. Mol Cell Endocrinol. 2010;327:1-12
8. Chang YJ, McCabe RT, Rennert H, Budarf ML, Sayegh R, Emanuel BS. et al. 3rd: The human "peripheral-type" benzodiazepine receptor: regional mapping of the gene and characterization of the receptor expressed from cDNA. DNA Cell Biol. 1992;11:471-80
9. Braestrup C, Squires RF. Specific benzodiazepine receptors in rat brain characterized by high-affinity (3H)diazepam binding. Proc Natl Acad Sci U S A. 1977;74:3805-9
10. Papadopoulos V. Peripheral-type benzodiazepine/diazepam binding inhibitor receptor: biological role in steroidogenic cell function. Endocr Rev. 1993;14:222-40
11. Li W, Hardwick MJ, Rosenthal D, Culty M, Papadopoulos V. Peripheral-type benzodiazepine receptor overexpression and knockdown in human breast cancer cells indicate its prominent role in tumor cell proliferation. Biochem Pharmacol. 2007;73:491-503
12. Canat X, Guillaumont A, Bouaboula M, Poinot-Chazel C, Derocq JM, Carayon P. et al. Peripheral benzodiazepine receptor modulation with phagocyte differentiation. Biochem Pharmacol. 1993;46:551-4
13. Verma A, Nye JS, Snyder SH. Porphyrins are endogenous ligands for the mitochondrial (peripheral-type) benzodiazepine receptor. Proc Natl Acad Sci U S A. 1987;84:2256-60
14. Garnier M, Boujrad N, Oke BO, Brown AS, Riond J, Ferrara P. et al. Diazepam binding inhibitor is a paracrine/autocrine regulator of Leydig cell proliferation and steroidogenesis: action via peripheral-type benzodiazepine receptor and independent mechanisms. Endocrinology. 1993;132:444-58
15. Maaser K, Grabowski P, Sutter AP, Hopfner M, Foss HD, Stein H. et al. Overexpression of the peripheral benzodiazepine receptor is a relevant prognostic factor in stage III colorectal cancer. Clin Cancer Res. 2002;8:3205-9
16. Katz Y, Ben-Baruch G, Kloog Y, Menczer J, Gavish M. Increased density of peripheral benzodiazepine-binding sites in ovarian carcinomas as compared with benign ovarian tumours and normal ovaries. Clin Sci (Lond). 1990;78:155-8
17. Batra S, Iosif CS. Peripheral benzodiazepine receptor in human endometrium and endometrial carcinoma. Anticancer Res. 2000;20:463-6
18. Batarseh A, Li J, Papadopoulos V. Protein kinase C epsilon regulation of translocator protein (18 kDa) Tspo gene expression is mediated through a MAPK pathway targeting STAT3 and c-Jun transcription factors. Biochemistry. 2010;49:4766-78
19. Hardwick M, Fertikh D, Culty M, Li H, Vidic B, Papadopoulos V. Peripheral-type benzodiazepine receptor (PBR) in human breast cancer: correlation of breast cancer cell aggressive phenotype with PBR expression, nuclear localization, and PBR-mediated cell proliferation and nuclear transport of cholesterol. Cancer Res. 1999;59:831-42
20. Beinlich A, Strohmeier R, Kaufmann M, Kuhl H. Specific binding of benzodiazepines to human breast cancer cell lines. Life Sci. 1999;65:2099-108
21. Landau M, Weizman A, Zoref-Shani E, Beery E, Wasseman L, Landau O. et al. Antiproliferative and differentiating effects of benzodiazepine receptor ligands on B16 melanoma cells. Biochem Pharmacol. 1998;56:1029-34
22. Neary JT, Jorgensen SL, Oracion AM, Bruce JH, Norenberg MD. Inhibition of growth factor-induced DNA synthesis in astrocytes by ligands of peripheral-type benzodiazepine receptors. Brain Res. 1995;675:27-30
23. Bruce JH, Ramirez AM, Lin L, Oracion A, Agarwal RP, Norenberg MD. Peripheral-type benzodiazepines inhibit proliferation of astrocytes in culture. Brain Res. 1991;564:167-70
24. Camins A, Diez-Fernandez C, Pujadas E, Camarasa J, Escubedo E. A new aspect of the antiproliferative action of peripheral-type benzodiazepine receptor ligands. Eur J Pharmacol. 1995;272:289-92
25. Maaser K, Hopfner M, Jansen A, Weisinger G, Gavish M, Kozikowski AP. et al. Specific ligands of the peripheral benzodiazepine receptor induce apoptosis and cell cycle arrest in human colorectal cancer cells. Br J Cancer. 2001;85:1771-80
26. Sutter AP, Maaser K, Barthel B, Scherubl H. Ligands of the peripheral benzodiazepine receptor induce apoptosis and cell cycle arrest in oesophageal cancer cells: involvement of the p38MAPK signalling pathway. Br J Cancer. 2003;89:564-72
27. Carmel I, Fares FA, Leschiner S, Scherubl H, Weisinger G, Gavish M. Peripheral-type benzodiazepine receptors in the regulation of proliferation of MCF-7 human breast carcinoma cell line. Biochem Pharmacol. 1999;58:273-8
28. Sutter AP, Maaser K, Hopfner M, Barthel B, Grabowski P, Faiss S. et al. Specific ligands of the peripheral benzodiazepine receptor induce apoptosis and cell cycle arrest in human esophageal cancer cells. Int J Cancer. 2002;102:318-27
29. Fafalios A, Akhavan A, Parwani AV, Bies RR, McHugh KJ, Pflug BR. Translocator protein blockade reduces prostate tumor growth. Clin Cancer Res. 2009;15:6177-84
30. Han Z, Slack RS, Li W. Papadopoulos V: Expression of peripheral benzodiazepine receptor (PBR) in human tumors: relationship to breast, colorectal, and prostate tumor progression. J Recept Signal Transduct Res. 2003;23:225-38
31. Vlodavsky E, Soustiel JF. Immunohistochemical expression of peripheral benzodiazepine receptors in human astrocytomas and its correlation with grade of malignancy, proliferation, apoptosis and survival. J Neurooncol. 2007;81:1-7
32. Huang ZL, Lin ZR, Xiao YR, Cao X, Zhu LC, Zeng MS. et al. High expression of TACC3 in esophageal squamous cell carcinoma correlates with poor prognosis. Oncotarget. 2015;6:6850-61
33. Hu L, Chen HY, Cai J, Yang GZ, Feng D, Zhai YX. et al. Upregulation of NETO2 expression correlates with tumor progression and poor prognosis in colorectal carcinoma. BMC Cancer. 2015;15:1006
34. Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell. 2016;166:21-45
35. Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265-73
36. Guo F, Parker Kerrigan BC, Yang D, Hu L, Shmulevich I, Sood AK, Xue F, Zhang W. Post-transcriptional regulatory network of epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions. J Hematol Oncol. 2014;7:19
37. Papadopoulos V, Lecanu L, Brown RC, Han Z, Yao ZX. Peripheral-type benzodiazepine receptor in neurosteroid biosynthesis, neuropathology and neurological disorders. Neuroscience. 2006;138:749-56
38. Corsi L, Avallone R, Geminiani E, Cosenza F, Venturini I, Baraldi M. Peripheral benzodiazepine receptors in potatoes (Solanum tuberosum). Biochem Biophys Res Commun. 2004;313:62-66
39. Kupczyk-Subotkowska L, Siahaan TJ, Basile AS, Friedman HS, Higgins PE, Song D. et al. Modulation of melphalan resistance in glioma cells with a peripheral benzodiazepine receptor ligand-melphalan conjugate. J Med Chem. 1997;40:1726-30
40. Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST. et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472-6
41. Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H. et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525-30
Author contact
![]() Corresponding authors: Prof. Wei-An Zeng, M.D. (Department of Anesthesiology, Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, 651 Dongfeng Road East, Guangzhou510060, Guangdong Province, China. Email: zengwasysu.edu.cn. Tel: +86-20-8734-3062) and Prof. Zhe-Sheng Wen, M.D. (Department of Thoracic Oncology, Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, 651 Dongfeng Road East, Guangzhou510060, Guangdong Province, China. Email: wenzhshorg.cn. Tel: +86-20-8734-3314).
Corresponding authors: Prof. Wei-An Zeng, M.D. (Department of Anesthesiology, Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, 651 Dongfeng Road East, Guangzhou510060, Guangdong Province, China. Email: zengwasysu.edu.cn. Tel: +86-20-8734-3062) and Prof. Zhe-Sheng Wen, M.D. (Department of Thoracic Oncology, Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, 651 Dongfeng Road East, Guangzhou510060, Guangdong Province, China. Email: wenzhshorg.cn. Tel: +86-20-8734-3314).

 Global reach, higher impact
Global reach, higher impact