3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(13):2604-2613. doi:10.7150/jca.16751 This issue Cite
Research Paper
Correlation between ERG Fusion Protein and Androgen Receptor Expression by Immunohistochemistry in Prostate, Possible Role in Diagnosis and Therapy
1. Translational Surgical Pathology, Laboratory of Pathology;
2. Urologic Oncology Branch, CCR, NCI, NIH, Bethesda, MD, USA.
Received 2016-7-6; Accepted 2016-8-18; Published 2017-8-5
Abstract
Background: Recent discovery of gene rearrangements have brought a new look to the molecular pathogenesis of cancer. Gene fusions occur in nearly 60% of prostate adenocarcinoma, being the TMPRSS2-ERG one of the most common. Evidence supports the role of ERG fusion in tumorigenesis, progression and invasion via effecting pathways such as WNT, MYC, uPA, PI3K/AKT/PTEN, RAS/RAF/MAPF, NKX3.1, GST-pi and androgen receptor (AR) mediated signaling. Most of the ERG fusions involve 5'-partners androgen responsive. Therefore, we aimed to evaluate AR and ERG fusion protein expression on prostate tissue to find clinicopathological applications and possible role in therapy.
Methods: One hundred three samples, including prostate core biopsies and radical prostatectomy specimens, were evaluated for ERG and AR expression by immunohistochemistry (IHC). ERG rearrangement was done by fluorescence in situ hybridization (FISH) on 11 randomly selected cases and correlated with IHC results.
Results: From the total of 103 samples, eight (8/103) were benign, fourteen (14/103) had atypical glands, two (2/103) had prostatic intraepithelial neoplasia (PIN), and seventy nine (79/103) showed prostate adenocarcinoma. Forty four (44/79) tumor cases were Gleason score (GS) 6-7 (lower GS), and thirty five (35/79) were GS of 8-10 (higher GS). ERG immunoreaction was observed in 27.8% (22/79) of the tumor cases, showing higher expression in those with lower GS (68.2%, 15/22) compared to higher GS (31.8%, 7/22). Neither benign glands nor PIN stained with ERG. AR expression was observed in 75% of benign samples, 78.5% of atypical glands, 100% of PIN, and in 87.3% of tumor cases with no significant difference based on GS. Co-expression of ERG and AR was evaluated on all the tumor samples. ERG+/AR+ was seen in 77.3% (17/22) of the ERG+ tumor cases, with higher frequency in lower GS (64.7%, 11/17) compared to those with higher GS (35.3%, 6/17). All but five corresponding ERG+ tumor samples were negative for AR. Only 5 samples were ERG-/AR- corresponding to adenocarcinoma GS of 6. Presence or absence of ERG rearrangement was confirmed by FISH and correlated with IHC results.
Conclusions: Characterization of ERG status by IHC in prostate tissue has an excellent correlation with FISH. It may also assist in diagnosis since none of the benign glands stained with ERG. Co-expression of ERG+/AR+ in prostate tumor by IHC may suggest gene fusion between ERG and a 5'-partner driven by androgen signaling such as TMPRSS2, which it could represent an important ancillary test for clinical management and development of new therapeutic targets.
Keywords: ERG Fusion Protein, Androgen Receptor Expression
Introduction
In the United States, prostate cancer is the most common primary neoplasm in males with an estimated of 180,890 new cases in 2016. Despite diagnostic and therapeutic advances in medicine, prostate cancer remains the second leading cause of cancer related deaths among males with 26,120 deaths expected to occur in 20161. In the last decade, progress has been made to search for molecular pathways and oncoproteins associated to the progression of prostate cancer, most of them driven by gene rearrangements and gene fusion, helping to identify new markers and modalities of treatment.
ETS-related gene (ERG) is a transcription factor from the Erythroblastosis virus E26 (ETS) oncogene family located on chromosome 212. The ETS family of proteins shows a wide variety of expression patterns in human tissues3. Members of the ETS family are known to be the key regulators of embryonic development, cell proliferation, differentiation, angiogenesis, inflammation, and apoptosis. The protein encoded by ERG is mainly expressed in the nucleus and contains and ETS DNA-binding domain and a PNT (pointed) domain which is implicated in the self-association of chimeric oncoproteins4. ERG is expressed in endothelial tissues, hematopoietic cells, kidney, and in the urogenital track5. This protein regulates hematopoiesis, and differentiation and maturation of megakaryocytic cells5. It is also required for platelet adhesion to the subendothelium, inducing vascular cell remodeling6. This gene is also involved in chromosomal translocations, resulting in different fusion gene products. One of the first findings was the identification of a nonrandom translocation of ERG from chromosome 21 to 8 in t(8; 21)(q22; q22) associated with acute myelogenous leukemia of the subgroup M2 (AML-M2)2. Later, other various contributions identified were the fusion of ERG with the TLS/FUS gene in acute myeloid leukemia and the EWS gene in Ewing's sarcomas7.
Discovery of the role of ERG in prostate cancer began in 2005 when Petrovics et al identified that ERG was frequently overexpressed in prostate cancer3. Subsequently, Tomlins et al, in 2005, discovered the ETS family transcription factor gene fusions, changing dramatically the field of solid tumor biology8. Recurrent gene fusions of the transmembrane protease serine 2 (TMPRSS2) gene to five members of the ETS family of transcription factor genes (ERG, ETV1, ETV4, ETV5, and ELK4) has been reported and confirmed in human prostate cancer, resulting in overexpression of normal or truncated ETS-proteins8-12.
The most common rearrangement in prostate cancer is the TMPRSS2-ERG fusion with a reported range from 15% to 80%, depending on the clinical cohorts investigated and methods of detection8-19. There is also a small percentage (<10%) of fusions that occur between ERG and other three additional androgen responsive 5' partners such as solute carrier family 45, member 3 (SLC45A3)19-21, homocysteine inducible endoplasmic reticulum protein with ubiquitin-like domain member 1 (HERPUD1)22 and N-myc downstream regulated gene 1 (NDRG1)15,19.
With the identification of ERG new studies emerged demonstrating its potential role as a prognostic indicator and as a marker for prostate cancer23-25. In 2006, Perner et al found that the presence of ERG rearrangements accompanied by 5′-ERG deletion has a significant correlation with higher tumor stage and the presence of metastatic disease involving pelvic lymph nodes13. Later in 2007, Demichelis et al, in a watchful waiting cohort of 111 patients, reported a significant association between the presence of a TMPRSS2-ERG fusion and prostate cancer-specific death, and a link between the presence of ERG alterations and higher Gleason score14.
Multiple molecular pathways have been implicated in the pathogenesis, development, differentiation, progression, invasion, and metastasis of prostate cancer. Several studies have revealed details about the mechanisms of ERG fusion in the development and progression of prostate cancer, such as: activation of c-Myc abrogating prostate epithelial differentiation26; regulation of the 15-hydroxy-prostaglandin dehydrogenase (HPGD) gene, a tumor suppressor, and prostaglandin catabolizing enzyme leading to altered levels of the HPGD and prostaglandin E2 (PGE2) as well as urokinase-type plasminogen activator (uPA)27. Also, ERG fusion has been involved in promoting cell proliferation through the PTEN/AKT/PIK3/mTOR pathway, and it has been suggested that its presence along with PTEN deficiency may cause the development of invasive cancer28. The androgen receptor (AR) plays a central role in any of those key-signaling pathways12.
Androgen is an important regulator of normal development and maintenance of the prostate gland29,30. Approximately 80-90% of prostate cancers are initially dependent on androgen requiring endocrine therapy directed toward the reduction of serum androgens and inhibition of AR31. AR is a DNA-binding nuclear transcription factor that regulates gene expression, which is maintained throughout prostate cancer progression, and persists in the majority of patients with hormone refractory disease25,32-34. As we know, in the majority of the prostate carcinomas there are fusion genes with oncogenic potential involving 5'-end elements composed of androgen regulated genes and the ETS gene transcription factor family25,35,36. Data has shown that overexpression of the androgen receptor does not give rise to hyperplastic lesions, but when combined with high levels of ERG, it promotes the development of a more poorly differentiated, invasive adenocarcinoma28. Some other studies have also demonstrated that androgen overstimulation of prostate cells cause development of fusion between TMPRSS2 and ERG genes18.
During the last decade, immunohistochemistry (IHC) has been introduced as a promising tool to detect ERG expression on the protein level. Furusato et al reported a highly specific mouse monoclonal ERG antibody performing a comprehensive evaluation of ERG protein expression using whole mount prostate sections from 132 prostate carcinoma cases, where ERG protein expression showed a strong concordance with ERG fusion transcripts by branched DNA assay or ERG rearrangement by fluorescence in situ hybridization (FISH) in selected specimens37. Similarly, other authors have provided a comprehensive data showing that virtually all ERG rearranged tumors exhibit ERG overexpression on the transcriptional and translational level19,38. To the best of our knowledge the evaluation of ERG fusion gene product and the AR at the expression level has not been well established in prostate specimens. Therefore, here we studied the correlation of ERG fusion product with AR expression by immunohistochemistry (IHC) in a wide range of prostate biopsies and prostatectomies in order to determine the clinical and pathological impact and possible therapeutic targets.
Materials and Methods
Samples
Formalin fixed paraffin embedded sections of a total of 116 prostate tissue specimens, including 102 biopsies and 14 radical prostatectomies, were retrieved from the archives of the Laboratory of Pathology at the NIH/NCI. These samples corresponded to 46 patients with a median age of 62 year-old. All these patients had been evaluated at the Urologic Oncology Branch of the National Cancer Institute, under approved protocol by the Institutional Review Board of the National Cancer Institute. Appropriate informed consents were obtained.
ERG and AR evaluation was performed on 103 samples from the total cases retrieved due to lack of material from thirteen (13/116) specimens. Tumors with various Gleason score (GS) were included in this study. Each tumor locus was evaluated separately.
Immunohistochemistry
Protein expression of ERG was performed in all the 103 cases by immunohistochemistry using a rabbit monoclonal antibody against human ERG (Clone EPR3864; Epitomics, Burlingame, CA). Formalin-fixed paraffin-embedded sections (5 μm) from core biopsies and whole tissue sections were deparaffinized in three Xylene baths and rehydrated in graded ethanol concentrations. Antigen retrieval was performed with Tris-EDTA buffer, pH 9.0, for 20 minutes under microwave and then allowed to cool down at room temperature for other 20 minutes. Endogenous peroxidase was blocked with 0.3% hydrogen peroxide in phosphate-buffered saline (PBS) for 15 minutes. Sections were then quenched with protein blocking buffer at room temperature for 30 minutes followed by one-hour incubation with a 1:100 dilution in PBS of the ERG antibody. For negative controls, sections were incubated in parallel with PBS instead of primary antibody. After rinsing, the sections were incubated with a peroxidase-labeled polymer conjugated to goat anti-mouse and goat anti-rabbit immunoglobulins as secondary antibody for 45 minutes (EnVision®+ Dual Link System-HRP Kit, DakoCytomation, Carpinteria, CA). The staining was visualized with 3, 3'-diaminobenzidine (DAB) as chromogen and slides were counterstained with hematoxylin, dehydrated and finally mounted. Vascular endothelial cells that were uniformly and strongly positive for ERG expression were used as the internal positive control.
Protein expression for AR was done similarly as the staining protocol for ERG. However, antigen retrieval was performed with citrate buffer, pH 6.0, for 20 minutes under microwave and then allowed to cool down at room temperature for other 20 minutes. Slides were incubated with mouse monoclonal antibody against human AR (1:50 dilution, Clone AR441, Dako, CA) for 30 minutes.
Both antibodies, ERG and AR, showed nuclear immunoreaction and it was subsequently graded using a semiquantitative scoring system as 0 (negative), 1 (weak), 2 (moderate) and 3 (strong) according to the intensity and percentage of stained glands (Figure 1). Appropriate internal and external controls were used. The histopathologic evaluation was performed by one experienced pathologist (MJM).
Fluorescence in situ hybridization
FISH analysis was performed on eleven (11) samples using dual-color intherphase break-apart probes for the 5' and 3' regions of ERG gene. Briefly, sections of 5 μm-thick, paraffin-embedded tissues were heated at 60ºC for 1-hour, then deparaffinized and rehydrated using xylene and graded ethanol, respectively. Antigen retrieval was performed with epitope retrieval solution (Catalog IW-1100, IHC-Tek, MD, USA) under electric steamer for 25 minutes and then allowed to cool down at room temperature for other 20 minutes. For optimum digestion, slides were incubated in pepsin solution (0.1mg/ml pepsin in 0.01 N HCl) at 37ºC for 30 minutes. Assessment was done every 3 to 5 minutes for optimal extent of tissue digestion. After rinsing in saline sodium citrate (SSC) buffer, dehydration was performed in increasing orders of ethanol and set for drying on heating block for 10 minutes. Subsequently, slides were incubated with the ERG Break-apart FISH probes (Empire genomics, Buffalo, NY) and covered with cover slip and sealed with rubber cement. After being denatured at 78ºC for 10 minutes they were incubated overnight at 37ºC in dark moist chamber. Later, the rubber cement was removed and soaked twice in the SSC buffer and then washed in pre-warmed 0.4xSSC/0.03% Tween 20 at 73ºC and followed by counter staining with 0.25µg/mL of 4,6-diamidino-2phenylindole-2-hydrochloride (DAPI). After incubating the slides at -20°C they were analyzed using fluorescein microscope. The hybridization was performed in low-stringency conditions with 50% formamide/2xSSC/10% dextran sulfate co-denaturation of the slide/probe, 1-hour hybridization at 37°C, followed by a 2 min wash in 0.4×SSC/0.3% Tween 20 at room temperature. Slides were counterstained with DAPI.
FISH signals were scored with a Zeiss epifluorescence microscope (Zeiss Axio Imager-2, Carl Zeiss Microimaging LLC, Thornwood, NY) equipped with a DAPI/FITC/Rhodamine single band pass filters (Semrock, Rochester, NY) using 40-60x objectives. In each case, a minimum of 100 cells were scored for the presence/absence of ERG gene rearrangement through deletion or split and it was considered positive if present in more than 50% of the nuclei. On each nucleus, the scoring was based on the number of unrearranged ERG loci (twinned red and green signals forming yellow signals), separated 5'-ERG sequences (labeled green) and separated 3'-ERG sequences (labeled red). Tumors were defined as positive for ERG rearrangement if a break occurs between the two probes giving two separated color signals (signal split) and/or deletion of the 5' probes (absence of one or more green signals), indirectly suggesting ERG fusion (TMPRSSS2-ERG).
Results
A total of 103 samples were evaluated for both antibodies, ERG and AR, by IHC. From these, eight (8/103) cases were benign, fourteen (14/103) had atypical glands, two (2/103) had prostatic intraepithelial neoplasia (PIN), and seventy nine (79/103) showed prostate adenocarcinoma. The cases with adenocarcinoma included twenty six (26/79) with GS of 6, eighteen (18/79) with GS of 7, twenty one (21/79) with GS of 8, and fourteen (14/79) with GS of 9 - 10. Table 1, 2 and 3 summarizes the results for ERG and AR expression by IHC.
ERG/AR expression by IHC staining prostate tissue from 103 samples.
| Samples (n = 103) | Expression | |||
|---|---|---|---|---|
| ERG+/AR+ (n = 19) | ERG+/AR- (n = 5) | ERG-/AR+ (n = 69) | ERG-/AR- (n = 10) | |
| Benign (n = 8) | 0 | 0 | 6 | 2 |
| PIN (n = 2) | 0 | 0 | 2 | 0 |
| Atypical glands (n = 14) | 2 | 0 | 9 | 3 |
| Adenocarcinoma (n = 79) | 17 | 5 | 52 | 5 |
Abbreviations: IHC, immunohistochemistry; n, total number of cases; ERG, transcription factor from the ETS family (Erythroblastosis virus E26 oncogene); AR, androgen receptor; PIN, prostatic intraepithelial neoplasia.
Correlation of ERG-positive tumor samples (based on GS) with AR expression by IHC.
| Adenocarcinoma samples, ERG+ (n = 22) | Expression | |
|---|---|---|
| ERG+/AR+ (n = 17, 77.3%) | ERG+/AR- (n = 5, 22.7%) | |
| GS of 6 (n = 9) | 7 (41.2%) | 2 (40%) |
| GS of 7 (n = 6) | 4 (23.5%) | 2 (40%) |
| GS of 8 (n = 4) | 3 (17.6%) | 1 (20%) |
| GS of 9 - 10 (n = 3) | 3 (17.6%) | 0 |
Abbreviations: IHC, immunohistochemistry; n, total number of cases; ERG, transcription factor from the ETS family (Erythroblastosis virus E26 oncogene); AR, androgen receptor; GS, Gleason score.
Correlation of ERG-negative tumor samples with AR expression by IHC.
| Adenocarcinoma samples, ERG- (n = 57) | Expression | |
|---|---|---|
| ERG-/AR+ (n = 52, 91.2%) | ERG-/AR- (n = 5, 8.7%) | |
| GS of 6 (n = 17) | 12 (23.1%) | 5 (100%) |
| GS of 7 (n = 12) | 12 (23.1%) | 0 |
| GS of 8 (n = 17) | 17 (32.7%) | 0 |
| GS of 9 - 10 (n = 11) | 11 (21.1%) | 0 |
Abbreviations: IHC, immunohistochemistry; n, total number of cases; ERG, transcription factor from the ETS family (Erythroblastosis virus E26 oncogene); AR, androgen receptor; GS, Gleason score.
ERG Immunohistochemistry
ERG immunoreaction was observed in 24 cases from the total of 103 samples, where two (2/103) had atypical glands and twenty two (22/103) were adenocarcinomas. A moderate to strong nuclear staining was present in most of the cases.
The expression of ERG in adenocarcinoma was 27.8% (22/79), and based on the GS it was more frequently seen on those tumors showing GS of 6 with 40.9% (9/22), followed by GS of 7 (27.3%, 6/22), GS of 8 (18.2%, 4/22), and GS of 9-10 (13.6%, 3/22). In general, ERG expression was 68.2% (15/22) in tumors with lower GS (6 and 7), and 31.8% (7/22) in those with higher GS (8 through 10). The majority of the tumor samples were ERG negative (72.1%, 57/79).
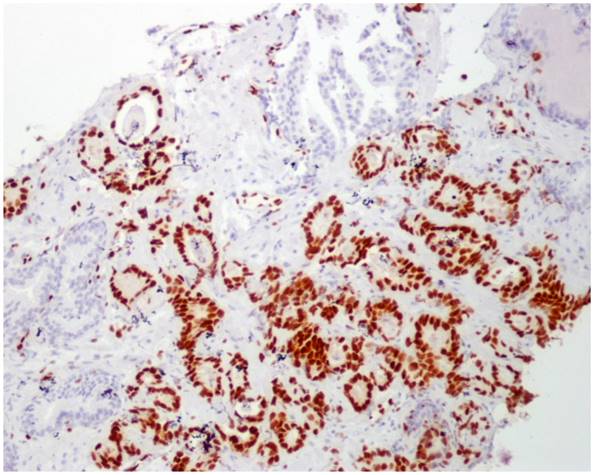
No immunoreaction to ERG was observed in PIN, benign samples, and in benign glands adjacent to adenocarcinoma in those tumor samples (Figure 2).
IHC staining strength for ERG (EPR3864) and AR
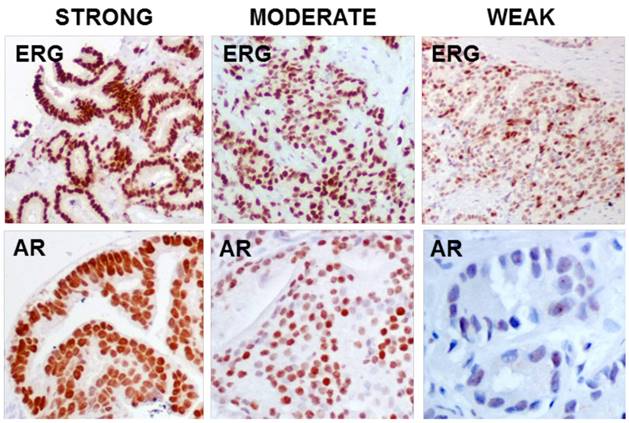
ERG immunoperoxidase staining showing the strong nuclear staining in the tumoral glands, while the benign glands have no nuclear immunoreaction to ERG (10X).
AR Immunohistochemistry
AR immunoreaction was observed in 88 cases from the total of 103 samples, where six (6/103) had benign glands, 2 (2/103) had PIN, eleven (11/103) had atypical glands and sixty nine (69/103) were adenocarcinomas. The strength of staining was mostly scored as 1 to 2 (weak to moderate). Few sections showed strong (score of 3) staining, and most of them were tumors with higher GS (8 through 10).
Among the total number of tumor cases, 87.3% (69/79) showed positive immunoreaction to AR antibody, and based on the GS it was as follows: 27.5% (19/69) on GS of 6, 23.2% (16/69) on GS of 7, 29% (20/69) on GS of 8, and 20.3% (14/69) on GS of 9-10. In general, AR expression was present in 50.7% (35/69) of the tumors with lower GS (6 and 7), and 49.3% (34/69) of those with higher GS (8 through 10), demonstrating no significant differences.
In addition, AR expression was seen in 75% (6/8) of benign samples, 78.6% (11/14) of atypical glands and in 100% (2/2) of PIN.
ERG/AR Immunohistochemistry Correlation
The majority of the cases showed ERG-/AR+ expression as follows: six benign samples (6/69), two samples with PIN (2/69), nine with atypical glands (9/69), and fifty-two cases (52/69) with prostate adenocarcinoma. Nineteen specimens were ERG+/AR+, being more frequently seen in samples with adenocarcinoma (89.5%, 17/19), and only in 2 cases with atypical glands (see Table 1).
Among the total number of ERG-positive (ERG+) tumor cases, 77.3% (17/22) showed expression for both, ERG and AR (ERG+/AR+); and based on the GS the co-expression was present as follows: 41.2% (7/17) on GS of 6, 23.5% (4/17) on GS of 7, 17.6% (3/17) on GS of 8, and 17.6% (3/17) on GS of 9-10. In general, ERG+/AR+ immunoreaction was more frequently seen in tumors with lower GS of 6-7 (64.7%, 11/17) than in those with higher GS of 8-10 (35.3%, 6/17) (see Table 2).
ERG+/AR- immunostaining was observed in a total of 5 tumor samples (5/22, 22.7%). Two of them were GS of 6, another 2 were GS of 7, and only 1 corresponded to GS of 8 (see Table 2).
From the total samples with ERG-negative (ERG-) tumors, 91.2% (52/57) showed ERG-/AR+ immunoreaction, and based on the GS it was as follows: 23.1% (12/52) on GS of 6, 23.1% (12/52) on GS of 7, 32.7% (17/52) on GS of 8, and 21.1% (11/52) on GS of 9-10. In general, ERG-/AR+ immunoreaction was seen in 46.2% (24/52) of the tumors with lower GS of 6-7, and 53.8% (28/52) in those tumors with higher GS of 8-10, suggesting no significant difference between GS (see Table 3).
ERG-/AR- was only seen in 9.7% (10/103) from the total samples evaluated. Among these, two (2/10) cases were benign, three (3/10) showed atypical glands, and 5 (5/10) were prostate adenocarcinoma with GS of 6.
Fluorescence in situ hybridization (FISH)
A break-apart assay was performed for the indirect detection of TMPRSS2-ERG fusion. One benign sample (ERG- by IHC) and 10 tumor sections (3 ERG-, 4 ERG+/AR+ and 3 ERG+/AR- by IHC) were included for the evaluation and correlation of ERG fusion by FISH. Benign glands in each sample were consistently negative for ERG fusion and were used as our internal control. Areas of tumor showing positive immunoreaction to ERG consistently demonstrated splitting or deletion of signal in more than 50% of the nuclei evaluated. Neither split of signal nor deletion was observed on the other 3 samples with ERG-negative tissue by IHC (1 benign and 2 tumor samples). ERG IHC overexpression (ERG+) was detected in all the cases with ERG rearrangement (7/7, 100%), while the other cases with no ERG rearrangement were also ERG-negative (ERG-) by IHC (4/4, 100%), indicating high sensitivity and specificity to detect ERG fusion protein expression by IHC. Detailed results are demonstrated in Table 4.
Correlation of ERG gene rearrangement by FISH with ERG expression by IHC on 11 randomly selected samples.
| ERG gene rearrangement by FISH (n = 11) | ERG expression by IHC | ||
|---|---|---|---|
| ERG+/AR+(n = 4) | ERG+/AR-(n = 3) | ERG- (n = 4) | |
| Positive (n = 7) | 4 | 3 | 0 |
| Negative (n = 4) | 0 | 0 | 4 |
Abbreviations: ERG, transcription factor from the ETS family (Erythroblastosis virus E26 oncogene); IHC, immunohistochemistry; FISH, fluorescence in situ hybridization; n, total number of cases; AR, androgen receptor.
Discussion
Discovery of the fusion genes in prostate cancer has brought a new perspective in our understanding of the pathophysiology of these tumors. The most common rearrangement in prostate cancer is the TMPRSS2-ERG fusion, which is androgen-driven and its prevalence has been confirmed to occur in 40% to 70% of prostate adenocarcinoma by several independent studies8-19. Although most studies have focused on the dominant rearrangement TMPRSS2-ERG fusion, a variety of other fusions and other androgen 5'-partners (SLC45A3, HERPUD1 and NDRG1) have been described, but appear to be less common8-22.
TMPRSS2-ERG fusion joins TMPRSS2 exons 1 or 2 usually to ERG exons 2, 3 or 4, which results in the activation of the ERG transcription factor. This fusion separates the 3'-ERG centromeric regions from the 5'-ERG telomeric ends; however, deletions of the 5' region can also occur and has been reported to be more common in prostatic small cell carcinomas with a more aggressive behavior39. The result of the fusion of ERG, either by translocation or deletion, is the production of a duplicated protein40. ERG protein expression has been detected in lymphocytes and endothelial cells of small vessels from benign and cancerous prostate tissue, regardless of the ERG rearrangement status. However, ERG rearrangements have never been detected by FISH in endothelial cells or lymphocytes19,41.
The recent literature, described ERG positivity in 26 - 90% of prostate carcinoma25,42-45. In our experience, ERG immunoreaction was observed in 27.8% (22/79) of the tumor cases, as described in the literature. Additionally, some authors described a link between the presence of ERG alterations and higher GS14,45, while others did not find any significant differences25,46. Our data reflects the opposite; we found that ERG expression is twofold more frequent in those tumors with lower GS (68.2%, 15/22) compared to those with higher GS (31.8%, 7/22). Similar data was observed in another study by Kron et al44. Also, endothelial cell nuclei have positive immunoreaction to ERG in all samples, and they were used as our internal control, keeping in consistency with previous studies. The ERG antibody used in our study demonstrated a very strong nuclear staining in most of the cases.
Interestingly, gene fusions have not been observed in benign prostate biopsies or in those normal prostatic glands adjacent to the tumor areas, benign prostatic hyperplasia (BPH) or even in atrophic glands through different molecular analysis13,46,47. However, Clark et al detected TMPRSS2-ERG fusions in 2 out of 31 BPH samples by reversed transcriptase-PCR (RT-PCR). But, when these two specimens were subjected to a repeated histopathological examination, no prostatic malignancy was identified17. ERG fusion has been consistently reported to be highly specific to the tumor tissue19,40. On the contrary, in previous studies, assessment of ERG protein expression by IHC in high grade PIN and benign glands that were adjacent to the ERG-positive tumor glands have been reported to be positively immunoreactive to the antibody19,48,49. Our experience demonstrated no expression of ERG in PIN, benign samples, and in those benign glands that were adjacent to the tumor areas. However, only 2 cases from a total of 14 with atypical glands demonstrated ERG+/AR+ immunoreaction. In both cases, the atypical glands were adjacent to ERG-positive, low GS (6 and 7), prostate adenocarcinoma. This finding may suggest the role of ERG gene fusion along with androgen stimulation in early development of prostate cancer. Also, identification of ERG immunoreaction in atypical glands should trigger the suspicion of the pathologist to evaluate the specimen thoroughly.
AR expression was observed in 75% of benign samples, 78.5% of atypical glands, 100% of PIN, and in 87.3% of tumor cases with no significant difference based on GS. However, if we correlate the expression of ERG and AR instead of analyzing each one by itself, our findings are similar to most of the published data3,12. For instance, Hermans et al evaluated the presence of ERG fusion in androgen-sensitive and androgen-independent xenografts by qPCR and FISH18. It was reported that in all tested androgen-sensitive cell lines, with the exception of one, the overexpression of ERG correlated with the presence of TMPRSS2-ERG fusion transcripts; however, in all androgen-independent xenografts no correlation has been observed. As we know TMPRSS2 is AR dependent, and androgen signaling induces proximity of the TMPRSS2 and ERG genomic loci, rendering fusion events most likely18. Moreover, in a recent study performed in prostatic small cell carcinomas, Wang et al demonstrated that there is an increased in the AR gene copy number and AR protein expression frequently associated with TMPRSS2-ERG fusion50. In our study, 77.3% (17/22) of the total ERG-positive tumors showed co-expression with AR (ERG+/AR+), being more frequently observed in lower GS (64.7%, 11/17) compared to those with higher GS (35.3%, 6/17). At the same time, four tumor cases randomly selected and expressing both proteins (ERG+/AR+) by IHC showed positive break-apart split indicating presence of ERG rearrangement (Figure 3), indirectly suggesting a fusion between an androgen driven 5'-partner and the 3'-ERG (most likely TMPRSS2-ERG). Although our study did not show statistical significance, based on our results it is possible to conclude that ERG+/AR+ expression by IHC is highly correlated to the TMRPSS2-ERG fusion at the expression level.
In vitro and in vivo models have demonstrated that ETS genes have an effect on tumor progression, but alone do not appear to be sufficient for transformation into cancer. For instance, Zong et al, demonstrated that overexpression of both AR and ERG promoted the development of poorly differentiated invasive adenocarcinomas28. However, in our experience the co-expression of ERG and AR was higher in those tumors with low GS compared to those with high GS.
Two cases, right and left panel, with prostate adenocarcinoma demonstrating ERG/AR expression by IHC (low and high magnification) correlated with their FISH results. The left panel shows positive expression of both proteins ERG and AR by IHC, and the right panel depicts the positive expression of ERG and lack of expression for AR. Both cases, show positive FISH analysis for ERG rearrangement with red and green signals split apart and spatially separated in different regions of the nuclei, indirectly suggesting TMRPSS2-ERG fusion.
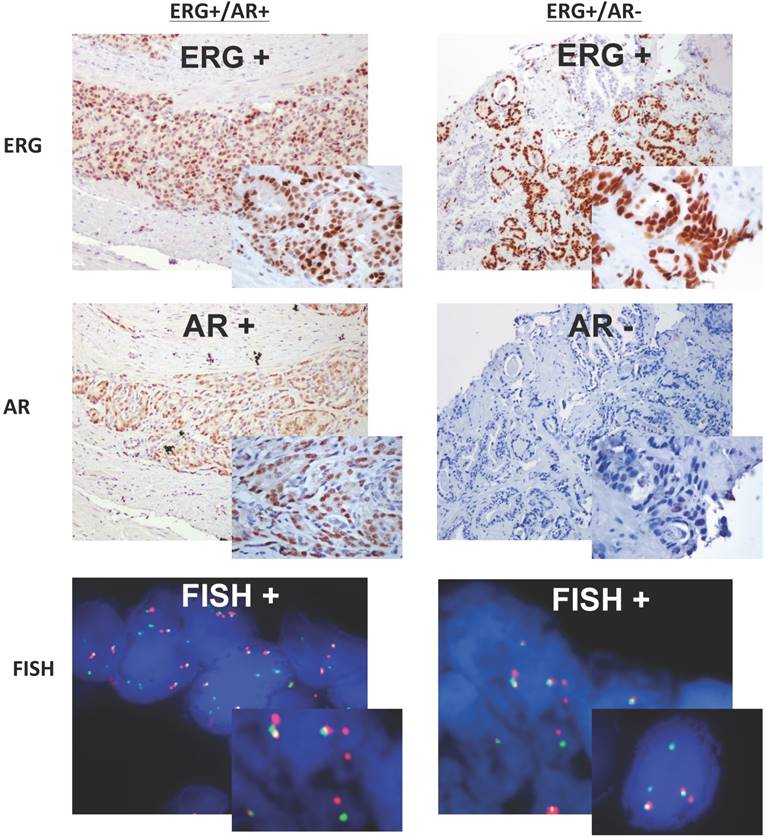
Furthermore, 22.7% (5/22) of the ERG-positive tumors were negative for AR (ERG+/AR-) by IHC. Two of them had a GS of 6, the other 2 had a GS of 7 and only 1 had a GS of 8. FISH analysis in three of these cases confirmed the presence of ERG-rearrangement (split signal) (Figure 3). One might speculate that the lack of expression of AR in those ERG-positive tumors could be explained by the fact that there are some 5' partners that are androgen-independent genes (DDX5 and C15orf21). These androgen-independent 5' partners are known to be ubiquitously expressed and found to be fused in frame with other ETS family members, leading to the expression of fusion proteins such as DDX5-ETV4 and C15orf21-ETV121,51. We might hypothesize that these 5' partner androgen-independent genes could be present in those tumors showing ERG+/AR- by IHC or we could think that these tumors may no longer be reliant on androgen signaling leading to a lack of staining for the AR. Further analysis and studies need to be done in order to clarify this issue.
Since the initial discovery of ETS fusions in prostate cancer there are multiple types of gene fusions described. On the basis of these discoveries, Rubin et al have developed a classification system composed by three categories: 1) fusions involving ETS gene family members (ERG, ETV1, ETV4, ETV5, and ELK4); 2) RAF kinase family fusions; and 3) SPINK1-positive prostate cancers12. In our study, the majority of the tumor samples were ERG-negative (72.1%, 57/79), and all of these but five cases were AR positive. Three (3) of these ERG-negative, randomly selected, tumor cases were confirmed to be negative for ERG rearrangement by FISH analysis. One could speculate that the ERG-/AR+ tumor cases could represent any of those categories described by Rubin et al, where the fusion gene involved corresponds to a RAF kinase family fusion, or a SPINK1 fusion, or still, it could belong to another gene of the ETS family member that is not expressing the ERG protein. More studies need to be done in order to find more answers to these findings.
Recent advances in next generation transcriptome sequencing have facilitated the discovery of the second category-RAF kinase gene fusions SCL45A3-BRAF, ESRP1-RAF1, and RAF1-ESRP1 in advanced prostate cancers. Although rare, detected in approximately 1 to 2% of prostate cancers, RAF kinase fusions represent the first “driver” fusions in prostate cancers that do not involve an ETS family member. The third category, SPINK1-positive prostate cancers, is included in the classification since the outlier expression of SPINK1 occurs in ETS-rearrangement-negative prostate cancers, and therefore defines a specific subclass of prostate cancer12.
In addition, we found that only 5 five cases of prostate adenocarcinoma with GS of 6 showed ERG-/AR- by IHC, which they could probably represent any of the less frequent previously described fusion genes (DDX5-ETV4 or C15orf21-ETV1) or even they could belong to any of those categories described by Rubin et al. Further analysis need to be done as well for these rare cases for better characterization, since they may have different behavior and clinical implications.
A limitation of our study is that due to the lack of material in the majority of the cases from our cohort, FISH analysis could not be done to confirm a significant correlation between the presence or absence of ERG rearrangement and the expression of ERG protein by IHC. Ideally, it would be to confirm the type of rearrangement or gene fusion involved by PCR or other methods in order to have a more clear evidence of the molecular pathways and corresponding levels of expression by IHC. Despite these limitations, we could provide data showing the prevalence of ERG positivity in 26.7% of the prostate carcinoma samples evaluated as well as we could show that ERG expression is inversely related to the GS. It is also expressed in a subset of atypical glands that are immediately adjacent to cancer. Additionally, there is a strong correlation between FISH and IHC for the ERG antibody with 100% sensitivity and specificity, which is similar to the reported data by Park et al where the sensitivity and the specificity of IHC is approximately 97% and 96%, respectively, a comparison done by FISH, PCR and IHC19.
In summary, based on our findings indicate that IHC may have an important function in determining truncated ERG protein expression as a result of ERG gene fusion and can be an efficient alternative to FISH. In addition, examination and correlation of ERG and AR have diagnostic significance and may be useful in assessing the biological character of the prostate cancer as well as selecting the best treatment with therapeutic molecular targets. Also, the co-expression of ERG/AR in prostate cancer could indirectly suggest that the patient harbors a gene fusion between ERG and a 5'-partner driven by androgen signaling such as TMPRSS2, SLC45A3 or NDRG1. Finally, is important to be aware of the expression of ERG protein in benign glands, atypical glands and/or high grade PIN since may warrant further workup to identify a possible adjacent tumor either in the same core or other cores. Further studies to establish the relevance of all these findings with the clinical outcome would be highly suggested.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7-30
2. Rao VN, Papas TS, Reddy ES. erg, a human ets-related gene on chromosome 21: alternative splicing, polyadenylation, and translation. Science. 1987;237:635-9
3. Petrovics G, Liu A, Shaheduzzaman S. et al. Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene. 2005;24:3847-52
4. Murakami K, Mavrothalassitis G, Bhat NK, Fisher RJ, Papas TS. Human ERG-2 protein is a phosphorylated DNA-binding protein-a distinct member of the ets family. Oncogene. 1993;8:1559-66
5. Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11-34
6. Han R, Pacifici M, Iwamoto M, Trojanowska M. Endothelial Erg expression is required for embryogenesis and vascular integrity. Organogenesis. 2015;11:75-86
7. Ichikawa H, Shimizu K, Hayashi Y, Ohki M. An RNA-binding protein gene, TLS/FUS, is fused to ERG in human myeloid leukemia with t(16;21) chromosomal translocation. Cancer Res. 1994;54:2865-8
8. Tomlins SA, Rhodes DR, Perner S. et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644-8
9. Tomlins SA, Mehra R, Rhodes DR. et al. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66:3396-400
10. Winnes M, Lissbrant E, Damber JE, Stenman G. Molecular genetic analyses of the TMPRSS2-ERG and TMPRSS2-ETV1 gene fusions in 50 cases of prostate cancer. Oncol Rep. 2007;17:1033-6
11. Soller MJ, Isaksson M, Elfving P, Soller W, Lundgren R, Panagopoulos I. Confirmation of the high frequency of the TMPRSS2/ERG fusion gene in prostate cancer. Genes Chromosomes Cancer. 2006;45:717-9
12. Rubin MA, Maher CA, Chinnaiyan AM. Common gene rearrangements in prostate cancer. J Clin Oncol. 2011;29:3659-68
13. Perner S, Demichelis F, Beroukhim R. et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337-41
14. Demichelis F, Fall K, Perner S. et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26:4596-9
15. Pflueger D, Rickman DS, Sboner A. et al. N-myc downstream regulated gene 1 (NDRG1) is fused to ERG in prostate cancer. Neoplasia. 2009;11:804-11
16. Attard G, Clark J, Ambroisine L. et al. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2008;27:253-63
17. Clark J, Merson S, Jhavar S. et al. Diversity of TMPRSS2-ERG fusion transcripts in the human prostate. Oncogene. 2007;26:2667-73
18. Hermans KG, van Marion R, van Dekken H, Jenster G, van Weerden WM, Trapman J. TMPRSS2:ERG fusion by translocation or interstitial deletion is highly relevant in androgen-dependent prostate cancer, but is bypassed in late-stage androgen receptor-negative prostate cancer. Cancer Res. 2006;66:10658-63
19. Park K, Tomlins SA, Mudaliar KM. et al. Antibody-based detection of ERG rearrangement-positive prostate cancer. Neoplasia. 2010;12:590-8
20. Esgueva R, Perner S, C JL. et al. Prevalence of TMPRSS2-ERG and SLC45A3-ERG gene fusions in a large prostatectomy cohort. Mod Pathol. 2010;23:539-46
21. Han B, Mehra R, Dhanasekaran SM. et al. A fluorescence in situ hybridization screen for E26 transformation-specific aberrations: identification of DDX5-ETV4 fusion protein in prostate cancer. Cancer Res. 2008;68:7629-37
22. Maher CA, Palanisamy N, Brenner JC. et al. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci U S A. 2009;106:12353-8
23. Carver BS, Tran J, Gopalan A. et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619-24
24. Mani RS, Iyer MK, Cao Q. et al. TMPRSS2-ERG-mediated feed-forward regulation of wild-type ERG in human prostate cancers. Cancer Res. 2011;71:5387-92
25. Kolar Z, Burdova A, Jamaspishvili T. et al. Relation of ETS transcription factor family member ERG, androgen receptor and topoisomerase 2beta expression to TMPRSS2-ERG fusion status in prostate cancer. Neoplasma. 2014;61:9-16
26. Sun C, Dobi A, Mohamed A. et al. TMPRSS2-ERG fusion, a common genomic alteration in prostate cancer activates C-MYC and abrogates prostate epithelial differentiation. Oncogene. 2008;27:5348-53
27. Mohamed AA, Tan SH, Sun C. et al. ERG oncogene modulates prostaglandin signaling in prostate cancer cells. Cancer Biol Ther. 2011;11:410-7
28. Zong Y, Xin L, Goldstein AS, Lawson DA, Teitell MA, Witte ON. ETS family transcription factors collaborate with alternative signaling pathways to induce carcinoma from adult murine prostate cells. Proc Natl Acad Sci U S A. 2009;106:12465-70
29. Roy AK, Lavrovsky Y, Song CS. et al. Regulation of androgen action. Vitam Horm. 1999;55:309-52
30. Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr Rev. 2004;25:276-308
31. Denis LJ, Griffiths K. Endocrine treatment in prostate cancer. Semin Surg Oncol. 2000;18:52-74
32. Hobisch A, Culig Z, Radmayr C, Bartsch G, Klocker H, Hittmair A. Androgen receptor status of lymph node metastases from prostate cancer. Prostate. 1996;28:129-35
33. Sadi MV, Walsh PC, Barrack ER. Immunohistochemical study of androgen receptors in metastatic prostate cancer. Comparison of receptor content and response to hormonal therapy. Cancer. 1991;67:3057-64
34. van der Kwast TH, Schalken J, Ruizeveld de Winter JA. et al. Androgen receptors in endocrine-therapy-resistant human prostate cancer. Int J Cancer. 1991;48:189-93
35. Haffner MC, Aryee MJ, Toubaji A. et al. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet. 2010;42:668-75
36. Bartek J, Hamerlik P, Lukas J. On the origin of prostate fusion oncogenes. Nat Genet. 2010;42:647-8
37. Furusato B, Tan SH, Young D. et al. ERG oncoprotein expression in prostate cancer: clonal progression of ERG-positive tumor cells and potential for ERG-based stratification. Prostate Cancer Prostatic Dis. 2010;13:228-37
38. van Leenders GJ, Boormans JL, Vissers CJ. et al. Antibody EPR3864 is specific for ERG genomic fusions in prostate cancer: implications for pathological practice. Mod Pathol. 2011;24:1128-38
39. Lotan TL, Gupta NS, Wang W. et al. ERG gene rearrangements are common in prostatic small cell carcinomas. Mod Pathol. 2011;24:820-8
40. Tomlins SA, Laxman B, Varambally S. et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10:177-88
41. Gavrilov D, Kenzior O, Evans M, Calaluce R, Folk WR. Expression of urokinase plasminogen activator and receptor in conjunction with the ets family and AP-1 complex transcription factors in high grade prostate cancers. Eur J Cancer. 2001;37:1033-40
42. Yu J, Yu J, Mani RS. et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17:443-54
43. Fernandez-Serra A, Rubio-Briones J, Garcia-Casado Z, Solsona E, Lopez-Guerrero JA. [Prostate cancer: the revolution of the fusion genes]. Actas Urol Esp. 2011;35:420-8
44. Kron K, Liu L, Trudel D. et al. Correlation of ERG expression and DNA methylation biomarkers with adverse clinicopathologic features of prostate cancer. Clin Cancer Res. 2012;18:2896-904
45. Bismar TA, Dolph M, Teng LH, Liu S, Donnelly B. ERG protein expression reflects hormonal treatment response and is associated with Gleason score and prostate cancer specific mortality. Eur J Cancer. 2012;48:538-46
46. Mosquera JM, Mehra R, Regan MM. et al. Prevalence of TMPRSS2-ERG fusion prostate cancer among men undergoing prostate biopsy in the United States. Clin Cancer Res. 2009;15:4706-11
47. De Marzo AM, Platz EA, Epstein JI. et al. A working group classification of focal prostate atrophy lesions. Am J Surg Pathol. 2006;30:1281-91
48. Lee SL, Yu D, Wang C. et al. ERG Expression in Prostate Needle Biopsy: Potential Diagnostic and Prognostic Implications. Appl Immunohistochem Mol Morphol. 2015;23:499-505
49. Yaskiv O, Zhang X, Simmerman K. et al. The utility of ERG/P63 double immunohistochemical staining in the diagnosis of limited cancer in prostate needle biopsies. Am J Surg Pathol. 2011;35:1062-8
50. Wang L, Williamson SR, Zhang S. et al. Increased androgen receptor gene copy number is associated with TMPRSS2-ERG rearrangement in prostatic small cell carcinoma. Mol Carcinog. 2015;54:900-7
51. Tomlins SA, Laxman B, Dhanasekaran SM. et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595-9
Author contact
![]() Corresponding author: Maria J. Merino, MD. mjmerinonih.gov 10 Center Drive, Room 3S235C, Bethesda, Maryland 20892, Phone 301-480-8483.
Corresponding author: Maria J. Merino, MD. mjmerinonih.gov 10 Center Drive, Room 3S235C, Bethesda, Maryland 20892, Phone 301-480-8483.

 Global reach, higher impact
Global reach, higher impact