3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(11):2142-2153. doi:10.7150/jca.18869 This issue Cite
Research Paper
Expression of CD133 in endometrial cancer cells and its implications
1. Department of Obstetrics and Gynecology, Buddhist Tzu Chi General Hospital; Hualien, Taiwan
2. Institute of Medical Sciences, Tzu Chi University; Hualien, Taiwan
3. Stem Cell Laboratory, Department of Research, Buddhist Tzu Chi General Hospital, Hualien, Taiwan
4. Department of Occupational medicine, Buddhist Tzu Chi General Hospital; Hualien, Taiwan
5. Department of Pediatrics, Buddhist Tzu Chi General Hospital; Hualien, Taiwan
Received 2016-12-22; Accepted 2017-5-9; Published 2017-7-15
Abstract
Cancer stem cells are an attractive therapeutic target for cancer. The present study examined stem cell characteristics of CD133+ cells isolated from endometrial cancer. Phenotypic characteristics, proliferation, migration, anchorage-independent growth, chemoresistance, gene expression profile and tumorigenicity of CD133+ tumor cells were assessed. Primary tumor exhibited immunoreactivity for CD133. Endometrial CD133+ tumor cells enhanced proliferation rate, colony formation, chemotaxis migration ability, and chemoresistance to cisplatin, paclitaxel, and doxorubicin than CD133- cells. CD133+ cells expressed more cancer stem cells markers such as EpCAM, aldehyde dehydrogenase 1 and insulin-like growth factor-1 receptor than CD133- cells. Moreover, CD133+ cells also increased expression of embryonic stem cell markers including oct4, nanog, sox2, and cmyc than CD133- cells. Finally, CD133+ tumor cells could generate xenograft but not CD133- tumor cells. CD133 and Ki67 were extensively expressed in the xenograft. In conclusion, endometrial CD133+ tumor cells displayed cancer stem cell characteristics and might represent a valuable tool for identifying endometrial cancer stem cells and hence a potential therapeutic target.
Keywords: Endometrial cancer, CD133, cancer stem cell, tumorigenesis, chemoresistance
Introduction
Endometrial cancer (EC) is the most common gynecologic pelvic malignancy in western countries, and its incidence is rising [1]. Endometrial cancer is usually diagnosed at an early stage due to initial presentation of abnormal vaginal bleeding. Approximately 80% of patients are diagnosed as having stage I disease; and 13%, as having stage 2 disease [2]. Although 88% of 5-year survival rate was noted in stage 1 disease, subgroups of patients have decreased survival rate [2]. Various prognostic factors, such as tumor grade, histologic type, high-grade lesions and deep invasion of the uterus have been identified [1]. The recurrence risk of endometrial cancer depends on the presence or absence of specific prognostic factors [3]. According to the above findings, novel target therapies need to be developed [4].
Cancer stem cells (CSCs) have been identified in leukemia and several solid tumors [5-7]. CSCs have self-renewal activity, produce progeny cells, constitute a small minority of neoplastic cells within a tumor, and possess the developmental potential for expression of multiple specific markers [8]. CSCs are also resistant to current cancer treatment, resulting in recurrence.
CD133 is a 120-kDa glycoprotein and a member of the family of pentaspan membrane glycoproteins [9]. CD133 can express in hematopoietic stem cells from adult blood, bone marrow and umbilical cord blood and on endothelial, neural and epithelial cells [10-12]. CD133 have been found in CSCs from prostate [13], lung [14], brain [15], colon [16, 17], and ovarian cancer [18] and acute lymphoblastic leukemia [19]. CD133 has been detected by its glycosylated epitope, AC133, consistent with the knowledge that the glycosylation state of a cell changes upon malignant transformation. CD133+ cells isolated from human endometrium show a low colony-forming unit capacity [20]. Whether endometrial cancer-isolated CD133+ cells possess CSC characteristics remains to be further clarified.
The present study was designed with the aim to determine CD133 expression in human endometrial cancer cells and to examine its self-renewal ability and chemoresistance. Gene expression and tumorigenicity of endometrial CD133+ tumor cells were also investigated.
Materials and methods
Patient characteristics
CD133+ and CD133- cells were isolated from tumor tissue specimen at the time of primary surgery from a 58-year-old woman with stage IA, grade 1 endometrioid adenocarcinoma of the endometrium (EC). She received laparoscopic staging surgery in 2010 and regular followed up without recurrence. Written informed consent to tumor tissue collection and its use for isolation of putative tumor stem cells was obtained from the patient according to the research protocol approved by the Research Ethics Committee of Buddhist Tzu Chi General Hospital (IRB 097-111).
Tissue collection, isolation and culture of CD133+ cancer cells
At the time of tissue collection, the tumor specimen was cut into two halves; one-half for confirming the pathological diagnosis, and the other half for isolation, purification, and culture of CD1333+ and CD133- cells. Within 30 min from surgery, tumors were mechanically and enzymatically dissociated with trypsin-EDTA for 15 min and then with collagenase I for 3 h at 37oC.
Cell suspensions were incubated with ammonium chloride (StemCell Technologies, Vancouver, British Colombia, Canada) for 10 min at 4oC to remove red blood cells. The CD45 MicroBeads (Miltenyi Biotec, Bergish Gladbach, Germany) was used for negative selection of non-hemopoietic CD45- cells. Then followed by positive selection of CD133+ cells were made using an AutoMACS platform (Miltenyi Biotec, Bergish Gladbach, Germany). The counterparts after CD133+ cells selection were designed as CD133- cells. The purity of selected CD133 population was assessed by flowcytometry of the expression of CD133 and CD45 (BD, PharMingen, Franklin Lakes, NJ, USA). Isolated CD133+ and CD133- cells were maintained in RPMI-1640 culture medium (Sigma, St Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS, Biological Industries, Kibbutz, Israel).
The specimen of normal endometrium (n=1) and the endometrium near the cancer part (n=1) were also dissected for the experiment and maintained in DMEM-low glucose medium (LG, Gibco, Grand Island, NY, USA) supplemented with 10% FBS. Endometrial cancer tissue was also sent to immunohistochemical (IHC) staining with CD133 (Miltenyi Biotec, Bergish Gladbach, Germany) and cytokeratin-7 (CK7, Bioss Inc., Woburn, MA, USA). Cell nuclei were stained with DAPI (4',6-diamidino-2-phenylindole, Sigma, St Louis, MO, USA).
Cell proliferation assay
Cell proliferation assays were performed by plating CD133+ and CD133- cells in T25 flasks with RPMI 1640 supplemented with 10% FBS at a density of 90,000 cells/ml. Cell counts were obtained on days 0, 3, 6, 8, and 10 using XTT assay (Roche, Mannheim, Germany) following the manufacturer's protocol. Each experiment was performed in triplicate.
Chemotaxis migration assay
CD133+, CD133- cancer cells, and RL95-2 cells (used as the control cell line) were seeded in the upper well of a 24-well transwell Boyden chamber (8 μm pore size; Costar, Corning Inc., Corning, NY, USA) and allowed to migrate towards cell-free media derived from ovarian stromal cells and follicular fluid (FF) placed in the bottom of wells. Cell growth medium was used as the control medium. Migration was assessed 48 hours later. The migrated cells were stained with crystal violet (Sigma-Aldrich, St Louis, MO, USA) and counted using bright-field microscopy. Each experiment was performed in triplicate.
RL95-2 cell is an endometrial cancer cell line obtained from BCRC (Bioresource Collection and Research Center, Hsinchu, Taiwan). The culture medium for RL95-2 was composed of 90% of a 1:1 ratio mixture of DMEM and Ham's F12 medium with 10 mM HEPES, 5 ug/ml bovine insulin, 2 g/L sodium bicarbonate, and 10% fetal bovine serum. Dr. Chu's lab provided primary ovarian stromal cells (obtained from ovarian specimen) and FF (obtained from oocyte retrieval). The ovarian stromal cell-conditioned medium was collected for 48 h by incubating ovarian stromal cells with high-glucose DMEM (Gibco, Grand Island, NY, USA) further supplemented with 10% FBS and 1% penicillin/streptomycin.
Anchorage-independent growth (AIG)
The soft agar assay for anchorage-independent growth was carried out on 5×105 CD133+ and CD133- cells in growth medium (4 ml) supplemented with 0.35% agarose and layered on a 5 ml base of 0.7% agarose. After 14 days, the cells were stained with 0.8 mM crystal violet (Sigma-Aldrich, St. Louis, MO, USA). The colonies larger than 30 μm were counted using bright-field microscopy. Each experiment was conducted in triplicate.
Immunocytochemistry
CD133+ and CD133- cells were sequentially treated with commercially available fixation and permeabilization media (Sigma, St. Louis, MO, USA), followed by incubation for 1 h at 4oC with monoclonal antibodies reacting against EpCAM (DakoCytomation, Glostrup, Denmark). FITC-conjugated goat anti-mouse antibodies were used as secondary reagents. Cell nuclei were stained with DAPI (Sigma, St. Louis, MO, USA).
Confocal microscopy
Serial formalin-fixed, paraffin-embedded endometrial tumor samples were deparaffinized with xylene, followed by absolute ethanol, 95% ethanol, and distilled water. CD133 was visualized with a PE or FITC conjugated mouse anti-CD133 antibody (1:30 dilution, Miltenyi Biotec, Bergish Gladbach, Germany). K7 was visualized with an FITC conjugated rabbit anti-cytokeratin 7 antibody (Bioss Inc., Woburn, MA, USA). Cell nuclei were stained with DAPI (Sigma). Imaging was performed using two-photon absorption fluorescence. A conventional confocal laser-scanning microscope (LSM 510 META, Zeiss, Jena, Germany) was used for two-photon absorption. Colocalization was done using SV1 software (Scientific Volume Imaging, Hilversum, Netherlands).
Quantitative RT-PCR
Total RNA was extracted using RNEasy® (Qiagen, Venlo, Netherlands) according to the manufacturer's instructions. Real-time PCR analysis of candidate genes (CSC-related genes: ALDH1, IGF-1R, PDGFRA, and PDGFRB; embryonic stem cell-related genes: oct4, nanog, sox2, and cmyc; primer sequences listed in Table 1) was performed following the methods in previous reports [21, 22]. Briefly, complementary DNA was synthesized using a SuperScript III One-Step RT-PCR kit (Invitrogen, Grand Island, NY, USA) and amplified by PCR with an AmpliTaq Gold Kit (Applied Biosystems, Foster City, CA, USA). Real-time PCRs were performed and monitored using FastStart Universal SYBR green Master (ROX, Roche, Indianapolis, IN, USA) and a quantitative real-time PCR detection system (ABI Step One Plus system, Applied Biosystems, Foster City, CA, USA). The gene products were analyzed with the GAPDH gene as a reference. The expression level of each target gene was then calculated as 2-ΔΔCt, as previously described [22]. Three readings of each experimental sample were obtained for each gene of interest and the experiments were repeated at least three times.
Sequence of primers used in real-time polymerase chain reaction
| Genes | Forward sequence (5'-3') | Reverse sequence (5'-3') | Product size (bp) |
|---|---|---|---|
| ALDH1 | TTG GAA TTT CCC GTT GGT TA | CTG TAG GCC CAT AAC CAG GA | 182 |
| IGFR1 | CCT CCT CGC ATC TCT TCT ACC TG | TGC TGG AGC CAT ACC CTG TG | 165 |
| PDGFRA | GTT CCA GCA GTT CCA CCT TC | ACC AAG TCC TCC TCT CAA CC | 212 |
| PDGFRB | CGTCAAGATGCTTAAATCCACAGC | TGATGATATAGATGGGTCCTCCTTTG | 145 |
| GAPDH | TCT CCT CTG ACT TCA ACA GCG AC | CCC TGT TGC TGT AGC CAA ATT C | 125 |
| Oct4 | CAGTGCCCGAAACCCACAC | GGAGACCCAGCAGCCTCAAAA | 161 |
| Nanog | AGTCCCAAAGGCAAACAACCCACTTC | TGCTGGAGGCTGAGGTATTTCTGTCTC | 161 |
| Sox2 | GGGAAATGGGAGGGGTGCAAAAGAGG | TTGCGTGAGTGTGGATGGGATTGGTG | 151 |
| C-myc | AAACACAAACTTGAACAGCTAC | ATTTGAGGCAGTTTACATTATGG | 188 |
| CD133 | CAGAAGGCATATGAATCC | CACCACATTTGTTACAGC | 181 |
In vitro sensitivity of endometrial CD133+/- cells to chemotherapeutic agents
Dissociated CD133+ and CD133- cells were cultured for 48 h in the presence of escalating concentrations of cisplatin (0-15 μM, Abiplatin, ABIC LTD., NETANYA, ISRAEL), doxorubicin (0-65 μM, Adriblastina, Pfizer, Kent, NJ, USA) and paclitaxel (0-80 nM, Formoxol, Yung Shin Pharm. IND., Co. Ltd., Taichung, Taiwan) as detailed in the figure legends. Cell viability was determined using trypan blue exclusion and counted using XTT assay and presented as the percentage of no-treatment control.
Xenograft experiment of endometrial CD133+/- cells
The procedures for animal experiments were carried out in adherence to the National Institutes Health Guide for the Care and Use of Laboratory Animals approved by The Animal Research and Care Committee of the Buddhist Tzu Chi General Hospital. Five-week-old Non-obese, diabetic-severe combined immune deficiency (NOD-SCID) (strain name: NOD.CB17-Prkdcscid/JTcu) mice were obtained from Tzu Chi University.
Isolated CD133+ and CD133- tumor cells (1.5 × 105 cells each 0.5 ml of medium per mouse, n = 3 in each group) were injected subcutaneously to examine the tumorigenesis. Mice were sacrificed 12 weeks after injection. The occurrence of tumor and tumor weight were measured. Tumor tissues were sent for histological examination. Hematoxylin and eosin staining and immunohistochemical analysis of CD133, CK7, and Ki67 was performed with staining with the specific antibody (BD Phamargen, San Diego, CA, USA). Tumor tissue was assessed at ×200 to ×400 total magnification.
Flowcytometry
The xenografted tumor were washed, acutely dissociated in DMEM and subject to enzymatic dissociation using 2 mM EDTA in PBS, washed with PBS containing 2% bovine serum albumin (BSA) and 0.1% sodium azide (Sigma, St Louis, MO, USA) and incubated with the anti-CD133 antibodies conjugated with FITC (BD, PharMingen, Franklin Lakes, NJ, USA). The cells were analyzed using a Becton Dickinson flow cytometer (Becton Dickinson, San Jose, CA, USA).
Statistical analysis
For data of in vitro and in vivo experiments, statistical comparisons among groups were performed using the Student's t-test or ANOVA with Bonferroni corrections. Differences between groups were considered statistically significant at p < 0.05. Data were expressed as mean ± SD.
Histology, histochemical, and molecular detection of CD133 in primary tumor samples. (A) H & E staining of the endometrial cancer specimen. Lower panel: magnification view of the upper panel. The pathology showed the characteristics of grade 1 endometrial cancer cells. Scale bar = 100 μm. (B) Immunofluorescence staining of endometrial tumor sections against CD133 (Red), cytokeratin 7 (CK7, green), and DAPI (blue) were analyzed and seen by confocal microscopy. Arrows indicate epithelium of endometrial glands. Scale bar = 50 μm. (B) Flowcytometry of dissociated endometrial tumor cells revealed the expression of CD133 and the pan-hematopoietic marker CD45. The percentage of CD133-expressing cells in endometrial cancer was shown. (C) Quantitative RT-PCR showed increased expression the mRNA of CD133 in tumor tissue. There was no expression of CD133 mRNA in normal endometrial tissues. ***p<0.001 compared with normal endometrium.
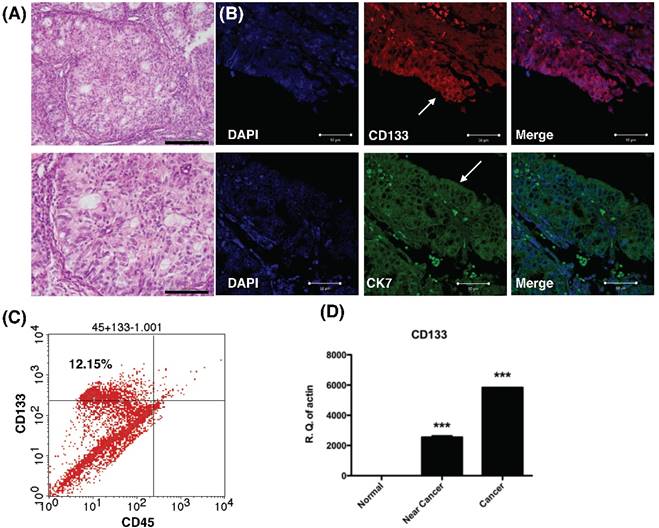
Results
CD133 expressed diffusely in primary endometrial tumors
To examine CD133 expression in primary EC, IHC, flow cytometry and qPCR of cancer tissue were performed. IHC revealed tumor sections diffusely stained by anti-CD133 antibodies and K7 (Fig. 1A). The tumor sample exhibited immunoreactivity with CD133 antibodies, which is in agreement with previously published reports on the phenotype of putative CSC in solid tumors [15]. Flow cytometry revealed 12.15% of CD133 expressed in freshly dissociated tumor cells (Fig. 1B). Finally, quantitative mRNA signals for CD133 were detected in dissociated tumor samples (cancer and near-cancer parts), but not expressed in normal endometrial tissue (p < 0.001, Fig. 1C). In summary, the results showed CD133 expression in EC epithelium.
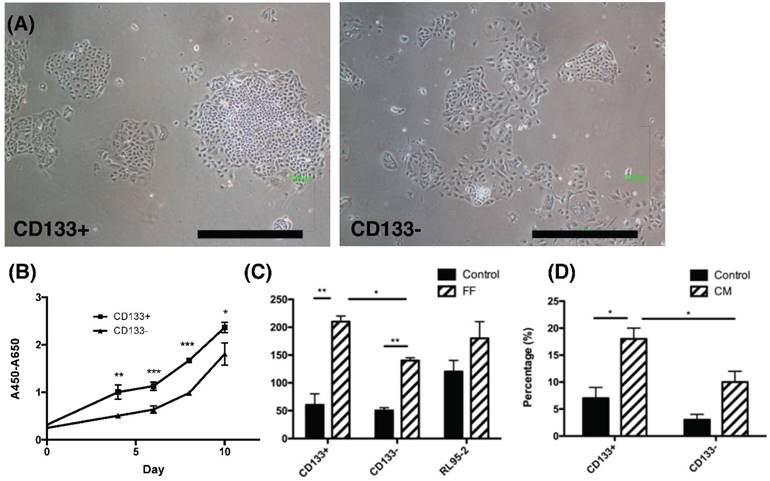
Enhanced proliferation and chemotaxis migration of CD133+ endometrial tumor cells
To investigate characteristics of CD133+ tumor cells, their proliferation and chemotaxis migration capabilities were compared with CD133- tumor cells. As seen in the morphology of CD133+/- tumor cells shown in Fig. 2A, tumor-derived CD133+ cells had ovoid-shaped cells (left panel) as same as CD133- cells (right panel). Moreover, CD133+ tumor cells showed a significantly higher proliferation rate when compared with CD133- cells (p<0.05, Fig. 2B).
Endometrial cancers are sometimes metastasis to the fallopian tube and ovary [23]. During ovulation, FF accompanied with oocytes extruded from a follicle, at the same time, ovarian stroma may increase exposure. Therefore, FF and condition medium (CM) from ovarian stromal cells were employed to investigate these chemotactic effects. Significantly enhanced migration of CD133+ cells towards FF compared with CD133- cells was observed (p < 0.05, Fig. 2C) but not in RL95-2 cells. In FF, the migration of tumor cells, whether CD133+/- cells, was better than that of the control group. In CM of ovarian stroma cells, the migration ability of CD133+ tumor cells was also significantly better than that of CD133- cells (p < 0.05, Fig. 2D). There was a significant enhancement in cell proliferation and migration of CD133+ cells than CD133- cells.
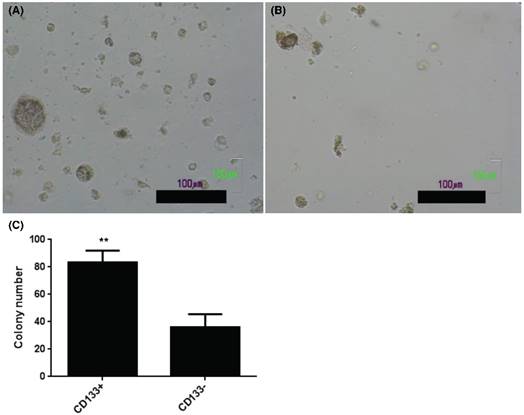
Enhancement in AIG of CD133+ than CD133- cells
The present results showed increase of AIG in soft agar colony assay, an important feature of tumor stem cells [24], in CD133+ (Fig. 3A) than CD133- cells (Fig. 3B). Quantification of colony number in soft agar showed more colonies noted in CD133+ cells than CD133- cells (Fig. 3C).
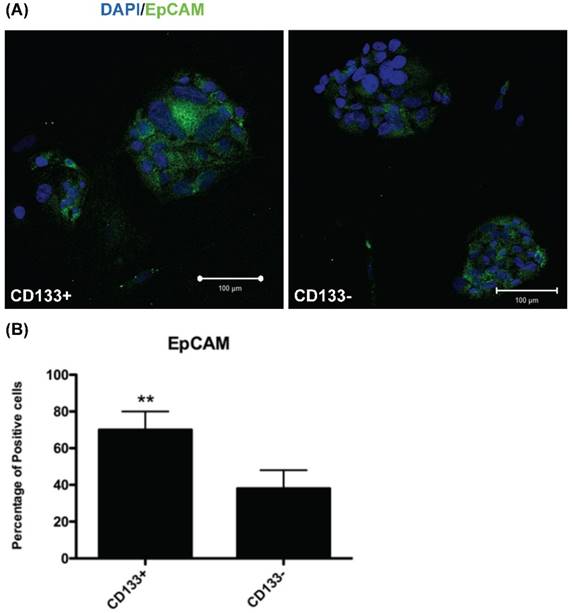
Increased EpCAM expression in CD133+ cells
To investigate EpCAM (expressed in cancer stem cells of breast and colon [17, 25]) expression in CD133+/- cells, IHC was used for evaluation. IHC revealed significantly more EpCAM-expressing cells in CD133+ cells than in CD133- cells (p < 0.01, Fig. 4).
The phenotypic characteristics, proliferation rate and migration ability of CD133+ and CD133- tumor cells. (A) The morphology of CD133+ (left panel) and CD133- (right panel) cells showed ovoid in shape. Scale bar = 1000 μm. (B) During the interval of 10 days, the proliferation rate of CD133+ cells was faster than CD133- cells. (C) After 48 hours of migration, the upper well of CD133+/- cells had more migrated cells towards follicular fluid (FF) than control medium. RL95-2 cells showed no difference between the control medium and FF. (D) After 48 hours of migration, CD133+/- tumor cells showed more migrated cells towards condition medium of ovarian stromal cells than control medium. Besides, more migrated CD133+ cells were noted than CD133- cells. *p < 0.05, ** p < 0.01.
CD133+ and CD133- tumor cells showed anchorage independent growth. Soft agar assay for colony formation of CD133+ (A) and CD133- cells (B) for 14 days and resulted colonies were photographed. (C) Numbers of colonies formed in soft agar by CD133+/- cells. Bars represent means ± standard deviation. Scale bar = 100 μm.
CD133+ cell increased EpCAM expression. (A) Immunofluorescence staining of CD133+/- cells using an antibody against EpCAM (green). DAPI staining (blue) shows the locations of the nuclei. (B) The percentage of EpCAM positive cells was counting and presented by the percentage of total cells. The experiments were done in triplicate. Bars represent means ± standard deviation. ** p < 0.01. Scale bar = 100 μm.
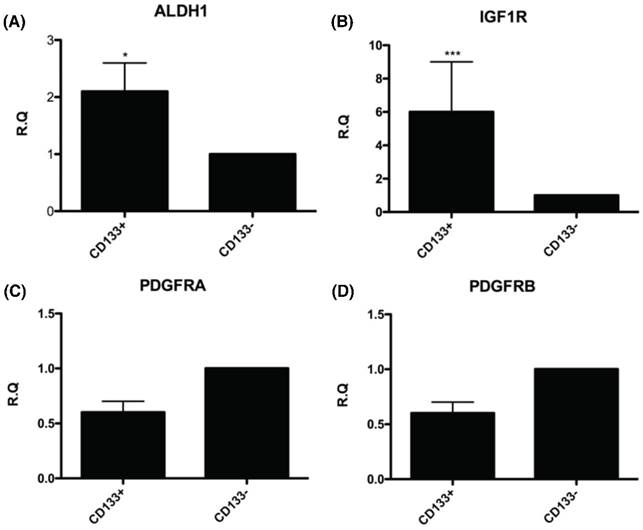
The CD133+ tumor cells increased expression of cancer stem cell (CSC)-related genes. Quantitative RT-PCR showed the expression of the CSC-related genes such as (A) Aldehyde dehydrogenase isoform 1 (ALDH1), (B) insulin-like growth factor 1 (IGF-1) receptor (IGF1R), (C) platelet-derived growth factor receptors α (PDGFRα), and (D) platelet-derived growth factor receptors β (PDGFRβ) in CD133+/- cells. *p < 0.05, ***p < .001.
Tumor-derived CD133+ cells expressed cancer and embryonic stem cell markers
To further understand the molecular events underlying CD133+ cells as CSC, the expression of genes related to the CSC such as ALDH1 (highly expressed in CSC) [26-30] and IGF-1R (hyperinsulinemia, risk factor of EC) [31] were examined by real-time PCR. The mRNA expression of these genes, including ALDH1 and IGF-1R were significantly upregulated in CD133+ cells compared with CD133- cells (Fig. 5A, B). Nevertheless, there was no significant difference in expression of gene-related migration toward CM and FF, PDGFRA and PDGFRB [32] between the two groups of cells (Fig. 5C, D).
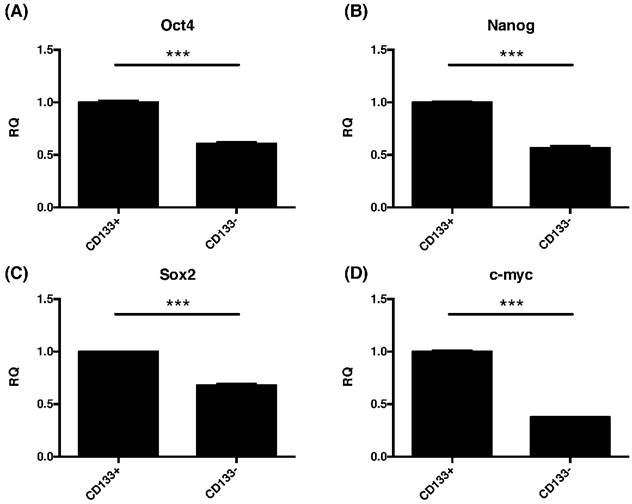
Due to common stemness regulators between embryonic and cancer stem cells [33], the pluripotent gene expressions of oct4, nanog, sox2, and cmyc in CD133+/- cells were investigated. This study found significantly increased the expression of oct4, nanog, sox2, and cmyc were noted in CD133+ cell than CD133- cells (p<0.001, Fig. 6).
Taken together, the findings revealed that CD133+ cells expressed more CSC- and ESC-related genes than CD133- cells.
Tumor-derived CD133+ cells had higher chemoresistance than CD133- cells
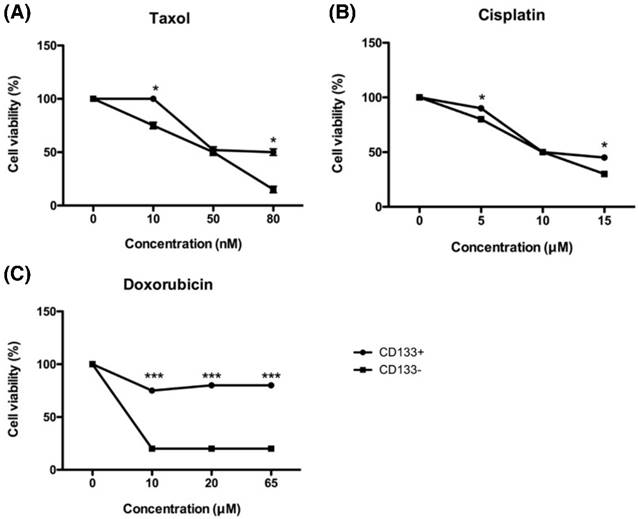
Tumor chemoresistance often comes from CSC with the characteristics of dormant and low proteasome activity [34]. To investigate the chemoresistance of EC-derived CD133+ and CD133- cells, both cells were exposed to increasing concentrations of chemotherapeutic drugs currently in use at the clinical setting, such as cisplatin, doxorubicin, and paclitaxel. As shown in Fig. 7, all chemo drugs induced less cell death in CD133+ cells than CD133- cells, indicating that CD133+ cells harbored more resistance to chemotherapeutic agents.
In Fig. 7C, doxorubicin was unable to kill CD133+ cells on the high concentration of 65 μM, but CD133- cells had already died at the same concentration. That is why taxane-based therapy now becoming the first choice in the first-line and salvage settings of endometrial cancer [35].
CD133+ tumor cells could generate xenograft tumor
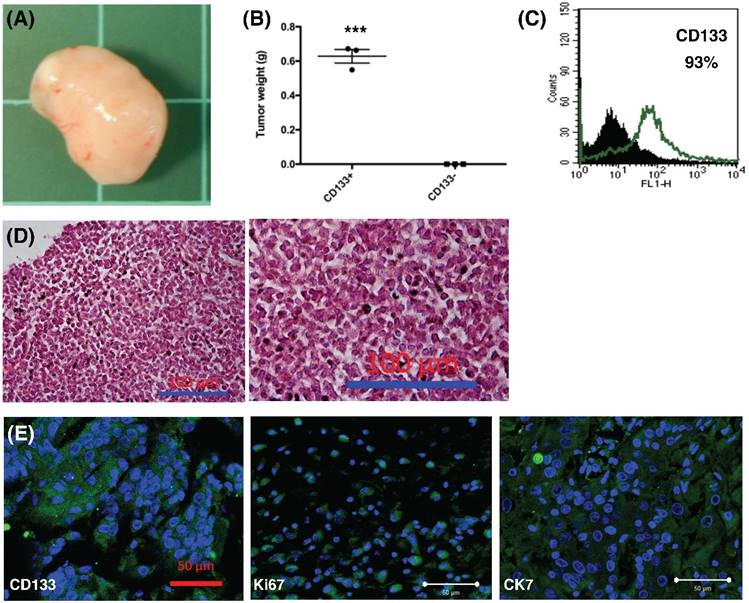
To further explore the role of CD133+ tumor cells, 1.5 × 105 cells were injected into the subcutaneous region of six female NOD-SCID mice. Three mice were injected with CD133+ cells and the other three mice were injected with CD133- cells. Tumor weight was evaluated after 12 weeks. CD133+ tumor cells could generate xenograft (3/3, Fig. 8A) but not CD133- cells (0/3) (p < 0.001, Fig. 8B). Moreover, the xenograft cells constantly expressed CD133 markers (93%, Fig. 8C). The expression of CD133, Ki67 (proliferation marker) and CK7 (specific to mullerian origin of epithelium, used for comparison) were examined using IHC. Tumor histology stained by H & E staining, as shown in Fig. 8D, revealed undifferentiated EC. The xenografted tumor intensively express CD133, Ki67, and CK7, suggesting tumor proliferative activities (Fig. 8E). Taken together, these results are consistent with the findings from the in vitro experiments and strongly support that CD133 as a cancer stem cell marker in EC.
Compare the findings in this study to those of the previous studies.
| Study | Year | Stem cell characteristics | Gene expression increased | Tumorigencity | Others |
|---|---|---|---|---|---|
| Rutella et al. [38] | 2009 | CD133+, serum free culture, sphere formation, chemoresistance, | Interleukin-8 CD44, CXCR4 | + | |
| Freil et al. [37] | 2010 | CD133+, 5-aza increased CD133 expression, decreased methylation after serial transplant of xenograft | + | ||
| Nakamura et al. [39] | 2010 | CD133+, chemoresistence, CD133+ EC worsen the prognosis | + | ||
| Nakamura et al. [44] | 2014 | CD133+, side population, high proliferation, colony forming activity, invasion | ABCG2, MMP | NA | |
| Guy et al. [52] | 2016 | CD133+, Progesterone treatment decreased CD133+ cells | NA | ||
| Shang et al. [53] | 2016 | CD133+, self-renewal, chemoresistance | Notch, EGFR | + | |
| Ding et al. (present study) | 2017 | CD133+, high proliferation, colony forming activity, chemoresistance | ALDH1, IGF1R, EpCAM, Oct4, sox2, nanog, c-myc | + | CD133 expression in normal part near cancer, increased migration ability |
NA: not applicable
Discussion
Novel target therapies are required for EC in addition to conventional therapy. Nevertheless, the current therapeutic strategies to target CSCs do not yield satisfactory results [36]. Several CSC markers such as CD133, CD44, and CD117 have been observed in EC and ovarian cancer [37-44], and the CSC population in EC has been reported to range from 1.3 to 62.6 % [38]. Other than EC, CSC has been reported at less than 1% of hematological malignancies, in which CSC have been well studied [36]. However, the discrepancy in EC population indicates that the targetable characteristics of CSCs need to be further confirmed. The present study investigated the various characteristics of the CD133+ population in EC. The CD133+ cells possessed more migratory capability than CD133- cells. Enhanced EpCAM expression was also observed in CD133+ tumor cells. More CD133+ cells expressed CSC-related genes like ALDH1 and IGF-1R and also embryonic stem cells genes oct4, nanog, sox2, and cmyc. CD133+ cells also harbored more chemoresistance and tumorigenesis capability. Enhanced CD133 and Ki67 expressions were noted in xenograft. We compared the current study with the previous reports in Table 2.
Criteria for defining CSC included self-renewal, restriction to a minority of the total tumor population, reproducible tumor phenotype, multipotent differentiation, and expression of unique cell surface markers, and consistent isolation [45]. CD133 may serve as a marker of tumor-initiating cells in a variety of human cancers, making it a good candidate for target therapy [9]. The present study found endometrial cancer cells mainly expressed the CD133+ cells, and CD133+ tumor cells were capable of self-renewal and forming larger colonies under appropriate culture conditions.
In this study and a previous report, proliferation rate was found to be more rapid in CD133+ cells than CD133- cells [38].
Chemotaxis is a basic biological process in which a cell migrates following the direction of a spatial cue. A form of a gradient of chemoattractants provided the spatial cue [46]. Migration ability is taken as one of the mechanisms behind metastasis; hence, it is essential to understand the capacity of CSCs to migrate to other tissues using in vitro migration assay. In this experiment, CD133+ cells showed a higher migration potential than CD133- cells under the stimulation of FF and CM of ovarian stromal cells. While the migration capacity of tumor cells is under the control of a large range of receptors and chemokines, further in vitro and in vivo studies are required to address this property fully. The gene expression of PDGFR, the receptor of PDGFA and PDGFB that are rich in FF and CM [32] were explored, but no significant difference in PDGFRA and PDGFRB expressions between CD133+ and CD133- cells was found. Regarding which gene affects the migration capability needs further investigation.
The CD133+ tumor cells increased expression of embryonic stem cell (ESC)-related genes. Quantitative RT-PCR revealed the expression of the ESC-related genes such as (A) Oct4, (B) nanog, (C) sox2, and (D) cmyc in CD133+/- tumor cells. ***p < 0.001.
In vitro chemosensitivity of tumor-derived CD133+ cells. The CD133+ (black circle) and CD133- (black square) tumor cells were maintained in growth medium either in the presence or absence of the indicated chemotherapeutic drugs [paclitaxel (A), cisplatin (B), and doxorubicin(C)] for 48 hours. After culturing, cells were harvested and counted by XTT assay, as detailed in Materials and Methods. *p < 0.05 and *** p < 0.001 compared with CD133- cells exposed to the same concentration. The experiments were done in triplicate. Bars represent means ± standard deviation.
In vivo xenograft experiment. (A) Formation of xenografted tumor in NOD/SCID mice transplanted with human CD133+ endometrial cancer cells. (B) After 12 weeks, all mice transplanted with CD133+ tumor cells generated tumor (n = 3), but not CD133- cells (n = 3). The mean tumor weight in CD133+ cells was 0.6g. ***p < 0.001. (C) After dissociation of xenografted tumor, flowcytometry showed 93% of the tumor cells expressed CD133 marker. (D) H & E staining of xenografted tumor, right panel: magnification view of the left panel. The pathology showed the characteristics of undifferentiated endometrial cancer cells. Scale bar = 100 μm. (E) Section of xenograft tumors were mounted and used for immunofluorescence. The tumor cells were positive for CD133, Ki67, and CK-7 (FITC). DAPI staining (blue) shows the locations of the nuclei. The experiments were done in triplicate. Scale bar = 50 μm.
When assessing the current CSC-associated gene expression profile of CD133+ cells compared with CD133- cells, CD133+ cells preferentially expressed EpCAM (highly expressed in serous uterine carcinoma) [47], aldh1 (highly expressed in CSC) [26-30] and IGF-1R (hyperinsulinemia, a risk factor of EC) [31]. Cancer usually shared the common stemness regulators of embryonic stem (ES) cells [33]. The CD133+ cells also increased expression the oct4, nanog, sox2, and cmyc, the same stemness genes in ES cells. These results indicated that gene expression profile of CD133+ cells was different from that of CD133- cells.
Therapeutic resistance is a key fundamental feature of CSCs. In the treatment of EC, cisplatin, paclitaxel and doxorubicin and radiation therapy are important [48]. The current results indicate that tumor-derived CD133+ cells have significant resistance to cisplatin, paclitaxel and doxorubicin in vitro when compared with their negative counterparts. The possibility of obtaining a virtually unlimited number of CD133-derived endometrial tumor cells has potential implications for the in vitro evaluation of drug sensitivity, and the availability of cells closely resembling the original tumor will allow the identification of novel drugs for individualized anticancer therapies.
The definition of CSC ultimately depends on in vivo xenograft experiments. It was found that CD133+ cells could grow tumors in all three xenografted NOD-SCID mice with 1.5 × 105 cells but not CD133- cells. This finding is consistent with previous reports on colon cancer [49] and glioblastoma multiforme [50]. CSCs were found in CD133+ fractions of EC. However, CD133- fractions could not form the xenograft in this experiment, which is inconsistent with previous findings [38]. The discrepancy may be caused by insufficient time to grow CD133- cells in mice (3 months in this experiment). The xenografted tumor could also enrich CD133+ fractions of tumor cells and recapitulated the original histology of the primary tumor [38].
The limitation of this study is as type I endometrial carcinomas account for more than 90% of endometrial cancers, the CD133 cells used in this study may not well represent the genetic profile, clinical presentation, and prognosis of the dominant type (carcinoma) of the endometrial cancers.
In conclusion, CD133+ cells can be isolated from human EC, can extensively propagate and migrate in vitro, exhibit chemoresistance and generate xenograft. CD133+ cells showed CSC markers such as EpCAM, aldh1, and IGF1R and ES cells markers including oct4, nanog, sox2, and cmyc. CD133 that exhibit stem cells characteristics [25] is a potentially useful antigen for the identification of EC stem cells and can be used for future target therapy [51].
Acknowledgements
The authors thank Dr. Yung-Hsiang Hsu for his assistance in preparing tumor pathology pictures.
Competing Interests
The authors have declared that no competing interests exist.
References
1. McConechy MK, Talhouk A, Leung S. et al. Endometrial carcinomas with POLE exonuclease domain mutations have a favorable prognosis. Clin Cancer Res. 2016;22(12):2865-73
2. Jemal A, Bray F, Center MM. et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69-90
3. Morrow CP, Bundy BN, Kurman RJ. et al. Relationship between surgical-pathological risk factors and outcome in clinical stage I and II carcinoma of the endometrium: a Gynecologic Oncology Group study. Gynecol Oncol. 1991;40(1):55-65
4. Sehouli J, Koensgen D, Oskay-Ozcelik G. et al. New aspects of adjuvant therapy in endometrial cancer: current standards and future directions. Crit Rev Oncol Hematol. 2008;67(3):204-212
5. Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3(7):730-737
6. Lapidot T, Sirard C, Vormoor J. et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645-648
7. Al-Hajj M, Wicha MS, Benito-Hernandez A. et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983-3988
8. Vescovi AL, Galli R, Reynolds BA. Brain tumour stem cells. Nat Rev Cancer. 2006;6(6):425-436
9. Mizrak D, Brittan M, Alison M. CD133: molecule of the moment. J Pathol. 2008;214(1):3-9
10. Yin AH, Miraglia S, Zanjani ED. et al. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90(12):5002-5012
11. Bonanno G, Mariotti A, Procoli A. et al. Human cord blood CD133+ cells immunoselected by a clinical-grade apparatus differentiate in vitro into endothelial- and cardiomyocyte-like cells. Transfusion. 2007;47(2):280-289
12. Bonanno G, Perillo A, Rutella S. et al. Clinical isolation and functional characterization of cord blood CD133+ hematopoietic progenitor cells. Transfusion. 2004;44(7):1087-1097
13. Collins AT, Berry PA, Hyde C. et al. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65(23):10946-10951
14. Eramo A, Lotti F, Sette G. et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15(3):504-514
15. Singh SK, Clarke ID, Terasaki M. et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63(18):5821-5828
16. Ricci-Vitiani L, Lombardi DG, Pilozzi E. et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445(7123):111-115
17. O'Brien CA, Pollett A, Gallinger S. et al. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445(7123):106-110
18. Ferrandina G, Bonanno G, Pierelli L. et al. Expression of CD133-1 and CD133-2 in ovarian cancer. Int J Gynecol Cancer. 2008;18(3):506-514
19. Cox CV, Diamanti P, Evely RS. et al. Expression of CD133 on leukemia-initiating cells in childhood ALL. Blood. 2009;113(14):3287-3296
20. Schwab KE, Hutchinson P, Gargett CE. Identification of surface markers for prospective isolation of human endometrial stromal colony-forming cells. Hum Reprod. 2008;23(4):934-943
21. Ding DC, Chu TY, Chiou SH. et al. Enhanced differentiation and clonogenicity of human endometrial polyp stem cells. Differentiation. 2011;81(3):172-180
22. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402-408
23. Dinulescu DM, Ince TA, Quade BJ. et al. Role of K-ras and Pten in the development of mouse models of endometriosis and endometrioid ovarian cancer. Nat Med. 2005;11(1):63-70
24. Brockman JA, Gupta RA, Dubois RN. Activation of PPARgamma leads to inhibition of anchorage-independent growth of human colorectal cancer cells. Gastroenterology. 1998;115(5):1049-1055
25. Al-Hajj M. Cancer stem cells and oncology therapeutics. Curr Opin Oncol. 2007;19(1):61-64
26. Dave B, Chang J. Treatment resistance in stem cells and breast cancer. J Mammary Gland Biol Neoplasia. 2009;14(1):79-82
27. Jiang F, Qiu Q, Khanna A. et al. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol Cancer Res. 2009;7(3):330-338
28. Rasheed ZA, Yang J, Wang Q. et al. Prognostic significance of tumorigenic cells with mesenchymal features in pancreatic adenocarcinoma. J Natl Cancer Inst. 2010;102(5):340-351
29. Su Y, Qiu Q, Zhang X. et al. Aldehyde dehydrogenase 1 A1-positive cell population is enriched in tumor-initiating cells and associated with progression of bladder cancer. Cancer Epidemiol Biomarkers Prev. 2010;19(2):327-337
30. Li T, Su Y, Mei Y. et al. ALDH1A1 is a marker for malignant prostate stem cells and predictor of prostate cancer patients' outcome. Lab Invest. 2010;90(2):234-244
31. Aizen D, Sarfstein R, Bruchim I. et al. Proliferative and signaling activities of insulin analogues in endometrial cancer cells. Mol Cell Endocrinol. 2015;406:27-39
32. McWilliam R, Leake RE, Coutts JR. Growth factors in human ovarian follicle fluid and growth factor receptors in granulosa-luteal cells. Int J Biol Markers. 1995;10(4):216-220
33. Hadjimichael C, Chanoumidou K, Papadopoulou N. et al. Common stemness regulators of embryonic and cancer stem cells. World J Stem Cells. 2015;7(9):1150-1184
34. Munakata K, Uemura M, Tanaka S. et al. Cancer stem-like properties in colorectal cancer cells with low proteasome activity. Clin Cancer Res. 2016
35. Moxley KM, McMeekin DS. Endometrial carcinoma: a review of chemotherapy, drug resistance, and the search for new agents. Oncologist. 2010;15(10):1026-1033
36. Cojoc M, Mabert K, Muders MH. et al. A role for cancer stem cells in therapy resistance: cellular and molecular mechanisms. Semin Cancer Biol. 2015;31:16-27
37. Friel AM, Zhang L, Curley MD. et al. Epigenetic regulation of CD133 and tumorigenicity of CD133 positive and negative endometrial cancer cells. Reprod Biol Endocrinol. 2010;8:147
38. Rutella S, Bonanno G, Procoli A. et al. Cells with characteristics of cancer stem/progenitor cells express the CD133 antigen in human endometrial tumors. Clin Cancer Res. 2009;15(13):4299-4311
39. Nakamura M, Kyo S, Zhang B. et al. Prognostic impact of CD133 expression as a tumor-initiating cell marker in endometrial cancer. Hum Pathol. 2010;41(11):1516-1529
40. Kryczek I, Liu S, Roh M. et al. Expression of aldehyde dehydrogenase and CD133 defines ovarian cancer stem cells. Int J Cancer. 2012;130(1):29-39
41. Baba T, Convery PA, Matsumura N. et al. Epigenetic regulation of CD133 and tumorigenicity of CD133+ ovarian cancer cells. Oncogene. 2009;28(2):209-218
42. Luo L, Zeng J, Liang B. et al. Ovarian cancer cells with the CD117 phenotype are highly tumorigenic and are related to chemotherapy outcome. Exp Mol Pathol. 2011;91(2):596-602
43. Steffensen KD, Alvero AB, Yang Y. et al. Prevalence of epithelial ovarian cancer stem cells correlates with recurrence in early-stage ovarian cancer. J Oncol. 2011;2011:620523
44. Nakamura M, Zhang X, Mizumoto Y. et al. Molecular characterization of CD133+ cancer stem-like cells in endometrial cancer. Int J Oncol. 2014;44(3):669-677
45. Clarke MF, Dick JE, Dirks PB. et al. Cancer stem cells-perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66(19):9339-9344
46. Wu D. Signaling mechanisms for regulation of chemotaxis. Cell Res. 2005;15(1):52-56
47. El-Sahwi K, Bellone S, Cocco E. et al. Overexpression of EpCAM in uterine serous papillary carcinoma: implications for EpCAM-specific immunotherapy with human monoclonal antibody adecatumumab (MT201). Mol Cancer Ther. 2010;9(1):57-66
48. Lester-Coll NH, Park HS, Rutter CE. et al. Who benefits from chemoradiation in stage III-IVA endometrial cancer? An analysis of the National Cancer Data Base. Gynecol Oncol. 2016;142(1):54-61
49. Vincent Z, Urakami K, Maruyama K. et al. CD133-positive cancer stem cells from Colo205 human colon adenocarcinoma cell line show resistance to chemotherapy and display a specific metabolomic profile. Genes Cancer. 2014;5(7-8):250-260
50. Kievit FM, Florczyk SJ, Leung MC. et al. Proliferation and enrichment of CD133(+) glioblastoma cancer stem cells on 3D chitosan-alginate scaffolds. Biomaterials. 2014;35(33):9137-9143
51. Pan Q, Li Q, Liu S. et al. Concise Review: Targeting Cancer Stem Cells Using Immunologic Approaches. Stem Cells. 2015;33(7):2085-2092
52. Guy MS, Qamar L, Behbakht K. et al. Progestin treatment decreases CD133+ cancer stem cell populations in endometrial cancer. Gynecol Oncol. 2016;140(3):518-526
53. Shang C, Lang B, Meng LR. Blocking NOTCH Pathway can Enhance the Effect of EGFR Inhibitor through Targeting CD133+ Endometrial Cancer Cells. Cancer Biol Ther. 2016 Epub
Author contact
![]() Corresponding author: Tang-Yuan Chu, M.D, Ph.D. Department of obstetrics and Gynecology, Buddhist Tzu Chi General Hospital, Tzu Chi University, Hualien, Taiwan. Tel: 886-3-8561825; Fax: 886-3-8577161; Email: hidrchucom
Corresponding author: Tang-Yuan Chu, M.D, Ph.D. Department of obstetrics and Gynecology, Buddhist Tzu Chi General Hospital, Tzu Chi University, Hualien, Taiwan. Tel: 886-3-8561825; Fax: 886-3-8577161; Email: hidrchucom

 Global reach, higher impact
Global reach, higher impact