3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(4):691-703. doi:10.7150/jca.17210 This issue Cite
Research Paper
Predictive Value of UGT1A1*28 Polymorphism In Irinotecan-based Chemotherapy
1. Department of Oncology, Second Affiliated Hospital of Xi'an Jiaotong University, Xi'an 710004, China;
2. Clinical Research Center, First Affiliated Hospital of Xi'an Jiaotong University, Xi'an 710061, China;
3. School of Medicine, Deakin University, Waurn Ponds, Victoria, Australia;
4. Department of Anesthesia, Second Affiliated Hospital of Xi'an Jiaotong University, Xi'an 710004, China.
* co-first authors
Received 2016-8-15; Accepted 2016-12-22; Published 2017-2-25
Abstract
The UGT1A1*28 polymorphism was suggested to be significantly connected with irinotecan-induced toxicity and response to chemotherapy. However, the results of previous studies are controversial. Hence we carried out a meta-analysis to investigate the effect of UGT1A1*28 polymorphism on severe diarrhea, neutropenia, and response of patients who had undergone irinotecan-based chemotherapy. The PubMed, Web of Science, Wanfang, and CNKI databases were searched for clinical trials assessing the association of UGT1A1*28 polymorphism with severe diarrhea, neutropenia, and response to irinotecan-based chemotherapy. The combined odds ratios (ORs) and 95% confidence intervals (CIs) were used to evaluate the relationship under a fixed- or random-effects model. Fifty-eight studies including 6087 patients with cancer were included. Our results showed that patients carrying the TA6/7 and TA7/7 genotypes had a greater prevalence of diarrhea and neutropenia than those with the TA6/6 genotype (TA6/7+TA7/7 vs. TA6/6: diarrhea, OR = 2.18, 95%CI = 1.68-2.83; neutropenia, OR = 2.15, 95%CI = 1.71-2.70), particularly patients with metastatic colorectal cancer. Stratified analysis showed that Asians with the TA6/7 and TA7/7 genotypes were more likely to have diarrhea and neutropenia, and Caucasians with the TA6/7 and TA7/7 genotypes were more likely to have neutropenia than other groups. However, patients with the TA6/7+TA7/7 genotypes showed a higher response than patients with TA6/6 genotype (OR = 1.20, 95%CI = 1.07-1.34), particularly Caucasians (OR = 1.23, 95%CI = 1.06-1.42) and patients with metastatic colorectal cancer (OR = 1.24, 95%CI = 1.05-1.48). Our data showed that the UGT1A1*28 polymorphism had a significant relationship with toxicity and response to irinotecan-based chemotherapy. This polymorphism may be useful as a monitoring index for cancer patients receiving irinotecan-based chemotherapy.
Keywords: UGT1A1*28, diarrhea, neutropenia, response.
Introduction
According to the estimation, there are probably 1,658,370 people suffer from cancer and 589,430 people die of cancer in the United States in 2015[1]. In China, the corresponding data were 4,292,000 and 2,814,000 in 2015, respectively, which means cancer is an urgent problem to be solved [2]. Several methods such as surgery, radiotherapy, and chemotherapy are widely applied for the clinical treatment of cancer. Irinotecan-based chemotherapy is one of the most used chemotherapies for patients with advanced gastric cancer, ovarian cancer, metastatic colorectal cancer, and other cancers [3-5]. Irinotecan, a camptothecin derivative, is mainly transported into liver by solute carriers and metabolized into ametabolite, SN-38, by a carboxylesterase [6]. In turn, SN-38 is glucuronidated by uridinediphosphate (UDP)-glucuronosyltransferases (UGTs) to an inactive form, SN-38G. Lower glucuronidation rates lead to higher SN-38 concentrations, resulting in irinotecan-induced severe toxicity [7]. Diarrhea and neutropenia are the most common side effects of irinotecan-based chemotherapy, limiting its application [8]. Recent studies have confirmed thatUDP UGT 1A1 play a vital role in the process of glucuronidation [9, 10].
The UGT1A1*28 polymorphism contains an extra TA repeat in the 5′-promoter region, whose mutant genotype is A(TA)7TAA (TA7/7) and has a wide genotype of A(TA)6TAA (TA6/6). Toffoli et al.[11] found that UGT1A1*28 TA7/TA7 genotype is related to a lower glucuronidation ratio. Previous studies investigated the relationship of UGT1A1*28 with neutropenia and diarrhea and have shown conflicting results. TA6/6 was reported to be a main predictive factor for diarrhea in a study of 56 advanced colorectal carcinoma (CRC) [12]. In contrast, some studies found that patients with the TA6/7 or TA7/7 genotypes are more inclined to suffer severe neutropenia and diarrhea [13-16]. However, no correlation was defined between the UGT1A1*28 polymorphism and neutropenia according to data from Hirata et al.[17] and Ferraldeschi et al.[18].
To clarify the predictive value of the UGT1A1*28 polymorphism in patients receiving irinotecan-based chemotherapy, we conducted this study to investigate the impact of the UGT1A1*28 polymorphism on tumor response and the common toxicities, diarrhea and neutropenia.
Materials and methods
Publication Search
Studies were selected by retrieving the Web of Science, PubMed, CNKI, and WanFang databases, up to June 2016. Similar keywords were used in different databases: “UGT1A1*28” and “diarrhea,” “UGT1A1*28” and “neutropenia,” “UGT1A1*28” and “response,” “UGT1A1*28” and “irinotecan,” “UGT1A1*28” and “CPT-11,” and related terms. No language restrictions were applied. All qualified studies were searched and a cross search was also used to identify the remaining relevant studies. When overlapping data exist in different reports, the most complete article was included. Disagreements between two authors will be settled by discussion and consensus.
Selection Criteria
Studies were included if they fulfilled the following criteria: (a) clinical trials; (b) evaluated the association of the UGT1A1*28 polymorphism with irinotecan-induced toxicities and chemotherapeutic effect; and (c) contained key information about the number of patients who have severe diarrhea, neutropenia and response to chemotherapy or not. Duplicate studies, review articles, letters, non-original studies, or case reports were excluded.
Data Extraction
Detailed information of included studies had been extracted and recorded in a standardized table by two reviewers. The following information was recorded: first author's surname, year of publication, ethnicity, cancer subtype, methods of mutation detection, number of patients with and without response, severe diarrhea and neutropenia, genotypes were extracted. If these data were not reported, items were marked “NR” (not reported).
Data Synthesis
This meta-analysis was conducted according to the PRISMA guidelines [19]. We used the Newcastle-Ottawa-Scale (NOS) to assess the qualities of including studies and calculated the combined odd ratios (ORs) and 95% confidence intervals (CIs) to evaluate the strength of relationship between the UGT1A1*28 polymorphism and irinotecan-induced diarrhea or neutropenia under the four models (TA6/7 vs. TA6/6, TA7/7 vs. TA6/6, TA6/7+TA7/7 vs. TA6/6, and TA7/7 vs. TA6/6+TA6/7) [20]. The association between tumor response and the UGT1A1*28 polymorphism was calculated only in the dominant model (TA6/7+TA7/7 vs. TA6/6). Pooled ORs were tested by the Z test, and a P value <0.05 was considered significant. Chi-square test and Q test were used to examine the heterogeneity among the studies. We also performed stratified analysis depending on tumor types (advanced gastric cancer, metastatic non-small cell lung cancer, metastatic colorectal cancer, or others), ethnicity (Asian, Caucasian or mixed people) and study design (retrospective or prospective study). Publication bias were determined by Egger's and Begg's tests [21, 22]. Specific methods are described in our pervious study [23]. A trim and fill method of adjusting for publication bias was carried out when the P value of Egger's test was less than 0.05 [24]. Trial sequential analysis (TSA) was conducted to calculate the required sample size to get a robust conclusion [20]. When P values of two-sided comparisons were less than 0.05, we considered the difference was significant. We performed all the statistical calculations by STATA 12.0 (StataCorp LP, College Station, TX, USA).
Results
Characteristics of the Studies Included
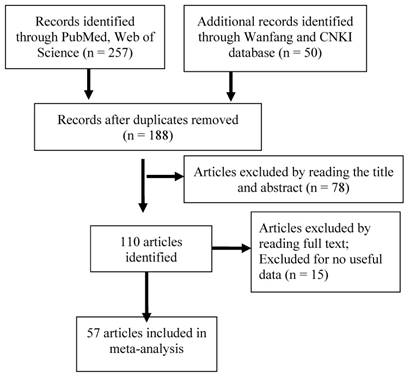
As shown in Figure 1, we performed the primary literature retrieval using the PubMed, Web of Science, Wanfang, and CNKI databases by the end of June 2016. First, 307 articles were included and 119 articles were excluded after searching for duplicates. Second, we read the titles and abstracts and excluded 78 articles because they were letters, case reports, reviews or reporting about other polymorphisms. Finally, after reading the full-text of all articles, 53 articles were excluded due to lacking of useful data or evaluation about other toxicities and 58 studies from 57 articles including 6087 patients with cancer were found to meet the inclusion criteria.
Flow diagram of included studies for the meta-analysis. CNKI = China National Knowledge Infrastructure
Among these studies, 16 studies investigated the associations in Caucasians [11-15, 18, 25-35], 40 in Asians [3, 9, 16, 17, 36-62], and two in mixed population or not reported [63, 64]. All studies were retrospective or prospective studies, including 29 metastatic colorectal cancer (mCRC) studies, five metastatic non-small cell lung cancer (mNSCLC), three advanced gastric cancer (GC) studies, two SCLC studies, and two advanced esophageal cancer studies and others. Table 1 summarized the basic information of the included studies.
Meta-Analysis of UGT1A1*28 Polymorphism and Severe Diarrhea
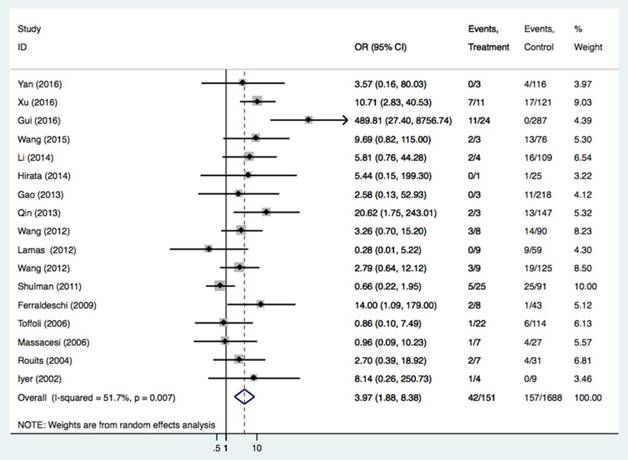
There were 44 studies of 4868 patients to evaluate the relationships between the UGT1A1*28 polymorphism and irinotecan-induced severe diarrhea. As shown in Table 2 and Figure 2, we found the UGT1A1*28 polymorphism was significantly related to severe diarrhea risk under all comparisons (TA 6/7 vs. TA6/6: OR = 1.56, 95%CI = 1.25-1.96; TA7/7 vs. TA6/6: OR = 3.97, 95%CI = 1.88-8.38; TA 7/7 vs. TA6/7+TA6/6: OR = 3.64, 95%CI = 2.01-6.58), regardless of the study design. By performing the subgroup analysis, we confirmed the relationship in the Asian group (TA6/7 vs. TA6/6: OR = 1.85, 95%CI = 1.37-2.50, P<0.001; TA7/7 vs. TA6/6: OR = 8.98, 95% CI = 5.21-15.47, P<0.001; TA6/7+TA7/7 vs. TA6/6: OR = 2.74, 95%CI = 2.21-3.40, P<0.001; TA 7/7 vs. TA6/6+TA6/7: OR = 8.64, 95%CI = 4.14-18.04, P<0.001) and in Caucasians (TA7/7 vs. TA6/6+TA6/7: OR = 1.62, 95%CI = 1.03-2.53). Stratified analysis according to cancer type was also carried out in this study. Individuals with mCRC carrying the TA7/7 or TA6/7 genotypes had a higher risk of getting diarrhea after irinotecan-based chemotherapy compared with the TA6/6 genotype (TA6/7 vs. TA6/6: OR = 1.60, 95%CI = 1.11-2.31, P = 0.011; TA7/7 vs. TA6/6: OR = 3.53, 95%CI = 1.54-8.09, P = 0.003). The same risk was also seen in SCLC patients (TA6/7+TA7/7 vs. TA6/6: OR = 3.95, 95%CI = 1.42-11.01, P = 0.009; TA7/7 vs. TA6/6+TA6/7: OR = 19.90, 95%CI = 2.57-154.1, P = 0.004).
Meta-Analysis of UGT1A1*28 Polymorphism and Severe Neutropenia
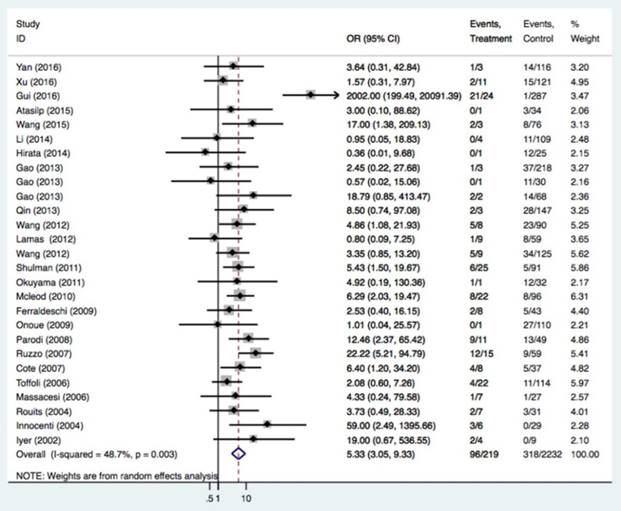
The relationships of the UGT1A1*28 polymorphism with irinotecan-induced severe neutropenia risk were investigated in 49 studies of 5232 patients. The UGT1A1*28 polymorphism was significantly related to an increased severe neutropenia incidence (Table 3 and Figure 3, TA 6/7 vs. TA6/6: OR = 1.71, 95%CI = 1.41-2.08; TA7/7 vs. TA6/6: OR = 5.34, 95%CI = 3.05-9.33; TA 7/7 vs. TA6/7+TA6/6: OR = 4.12, 95%CI = 2.36-7.20). Caucasians and Asians with at least one TA7 allele had a higher risk of neutropenia (Caucasians: TA6/7 or TA7/7 vs. TA6/6: OR = 1.84 and 5.67; Asians: TA6/7 or TA7/7 vs. TA6/6: OR = 1.56 and 4.77). In the analysis stratified by cancer type and study design, an association was also found in retrospective and prospective designs, with mCRC patients having the TA7/7 and TA6/7 genotypes (TA6/7 or TA7/7 vs. TA6/6: OR = 1.76 and 5.07) and solid tumor patients with the TA7/7 genotype (TA7/7 vs. TA6/6 or TA6/6+6/7: OR = 7.66 and 6.68).
Characteristics of the Studies Included in the Meta-Analysis
| Study | Year | Study design | Race | Cancer | Mutation detection methods | Regimen | IRI dose (mg/m2)/schedule | Population source | No. of patients | Age | ECOG | NOS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yan8 | 2016 | R | Asian | mixed tumors | PCR-Sanger sequence | FOLFIRI, IRI + CDDP, IRI + BEV | 125, 150 or 180 mg/m2 | S | 157 | 53 | NR | 8 |
| Xu64 | 2016 | R | Asian | mCRC | Direct Sequencing | FOLFIRI, IRI+CAP | 150mg/m2, every 2 or 3 weeks | S | 183 | NR | 0-1 | 9 |
| Gui65 | 2016 | R | Asian | mCRC | SPR | FOLFIRI, IFL | 180mg/m2, every 2 or 3 weeks | S | 384 | NR | 0-2 | 8 |
| Wang5 | 2016 | P | Asian | Advanced GC | SPR | IRI+CDDP | 80 or 125mg/m2 | S | 40 | 54 | 0-2 | 8 |
| Li4 | 2016 | P | Asian | mCRC | SPR | FOLFIRI, mCapeIRI, IRI | NR | M | 160 | 50 | 0-2 | 9 |
| Yang63 | 2015 | R | Asian | pancreatic or biliary tract cancer | Direct Sequencing | FOLFIRI, IRI alone | 180mg/m2, biweekly | S | 48 | 56.2 | 0-1 | 7 |
| Peng60 | 2015 | P | Asian | mCRC | Sequencing | FOLFIRI; mFOLFIRI | 180mg/m2, biweekly | S | 208 | 59.8 | 0-3 | 7 |
| Wu59 | 2015 | P | Asian | Advanced esophageal cancer | NR | IRI+PLA | 180mg/m2, every 3weeks | S | 42 | 55 | 0-2 | 7 |
| Xu3 | 2015 | NR | Asian | advanced OC | PYRS | IRI+CDDP | 60mg/m2 IRI (d1, 8) every 3 weeks | S | 89 | 48 | NR | 7 |
| Xiao9 | 2015 | R | Asian | SCLC | PYRS | IRI+CDDP/CBP/LOB | 60 mg/m2 (d1,8,15), every 4 weeks; 85mg/m2 (d1,8), every 3 weeks | S | 67 | NR | 0-2 | 8 |
| Shi61 | 2015 | P | Asian | SCLC | Direct Sequencing | IRI+CDDP | 65mg/m2 (d1, 8) | M | 30 | 59 | 0-2 | 8 |
| Atasilp10 | 2015 | R | Asian | mCRC | PYRS | FOLFIRI, FOLFIRI+CET, FOLFIRI+BEV, mFOLFIRI, IRI alone, IRI+CET/CAP | 180mg/m2, biweekly; 100mg/m2 | S | 44 | 6 | 0-2 | 7 |
| Chen62 | 2015 | P | Asian | mNSCLC | Sequencing | IRI+DDP | 100mg/m2, every 3 weeks | S | 86 | 63 | 0-2 | 8 |
| Wang35 | 2015 | P | Asian | mCRC | Sequencing | NR | NR | S | 111 | NR | 0-1 | 7 |
| Li54 | 2014 | R | Asian | mCRC | PYRS | FOLFIRI, IRI + CET/BEV, IRI + RAL, IRI+ CAP | 180 mg/m2, every 2 or 3 weeks | S | 167 | 50 | 0-2 | 8 |
| Hirata17 | 2014 | P | Asian | mCRC | SPR | FOLFIRI | 150mg/m2, biweekly | M | 34 | 62 | 0-2 | 7 |
| Zhao55 | 2014 | P | Asian | SCLC | Direct sequencing | IRI+CDDP | 60mg/m2 (d1,8,15), every 3 weeks | S | 34 | 49 | 0-2 | 8 |
| Song56 | 2014 | P | Asian | Advanced OC | NR | IRI+PLA | 60mg/m2 (d1,8), every 3 weeks | S | 89 | 48 | NR | 8 |
| Zhang57 | 2014 | P | Asian | mCRC | Sequencing | FOLFIRI, XELIRI, IRIR | 180mg/m2, biweekly; 200mg/m2, every 3weeks | S | 102 | 55 | NR | 8 |
| Xu53 | 2014 | P | Asian | GC | Sequencing | NR | NR | S | 67 | 62.7 | 0-2 | 8 |
| Zhou58 | 2014 | P | Asian | mCRC | SPR | IRI+5-FU/TMZ/CAP | 180mg/m2 | S | 82 | 59 | NR | 8 |
| Zhou52 | 2013 | P | Asian | gastrointestinal cancer | Direct Sequencing | FOLFIRI | 180mg/m2, biweekly | S | 94 | 58.5 | 0-1 | 8 |
| Hirasawa50 | 2013 | R | Asian | cervical or ovarian cancer | Invader assay | IRI+CDDP, IRI alone | 60 or 100mg/m2 (d1, 8, 15), every 4 weeks | S | 53 | 48 | NR | 7 |
| Gao48 | 2013 | R | Asian | mCRC | Sanger Sequencing | FOLFIRI, IRI alone or IRI+CET/CAP | 180mg/m2 | S | 276 | 55 | NR | 7 |
| Gao49 | 2013 | R | Asian | advanced GC | Sanger Sequencing | IRI+CDDP, FOLFIRI, IRI alone, IRI+CET | 180mg/m2 | S | 42 | 53 | NR | 7 |
| Gao49 | 2013 | R | Asian | advanced esophageal cancer | Sanger Sequencing | IRI+CDDP, FOLFIRI, IRI alone, IRI+CET | 130mg/m2; 180mg/m2 | S | 91 | 54 | NR | 7 |
| Qin51 | 2013 | R | Asian | advanced gastrointestinal carcinoma | Sequencing | IRI, IRI+CDDP, IRI+5-FU | NR | S | 183 | NR | NR | 7 |
| Wang45 | 2012 | NR | Asian | mCRC | Direct Sequencing | FOLFIRI, IRI+LEU | 180mg/m2, biweekly; 125mg/m2 (d1, 8, 15, 22), every 6 weeks | S | 130 | 52 | 0-2 | 7 |
| Zhang46 | 2012 | P | Asian | mCRC | Direct Sequencing | FOLFIRI, IRI+LEU | 180mg/m2, biweekly; 125mg/m2 (d1, 8, 15, 22), every 6 weeks | S | 56 | 55.5 | NR | 8 |
| Lamas34 | 2012 | R | Caucasian | mCRC | Fluorescent DNA length fragment analysis | FOLFIRI, FOLFIRI-CET, FOLFIRI-BEV, IRI+CET | 180mg/m2, biweekly | S | 101 | 67 | 0-2 | 7 |
| Wang47 | 2012 | P | Asian | mCRC | Sequencing | IFL, FOLFIRI | 125mg/m2, weekly;180mg/m2,biweekly | S | 180 | 54 | 0-2 | 7 |
| Shulman33 | 2011 | R | Caucasian | mCRC | SPR | FOLFIRI, IFL, TEGAFIRI, XELIRI | U | M | 214 | 63.1 | NR | 8 |
| Okuyama43 | 2011 | P | Asian | mCRC | SPR | FOLFIRI | 150mg/m2 | S | 39 | 64 | 0-2 | 7 |
| Nakamura42 | 2011 | P | Asian | mNSCLC | Polyacrylamide gel electrophoresis | IRI+PAC, IRI+GEM | 50mg/m2 (d1, 8 and 15), every 4 weeks; 100mg/m2 (d1 and 8), every 3 weeks | S | 77 | NR | 0-1 | 8 |
| Park44 | 2011 | P | Asian | mGC | Sequencing | S-1+IRI+OXA | 150mg/m2, every 3 weeks | S | 44 | 54 | 0-2 | 7 |
| Mcleod32 | 2010 | P | Caucasian | mCRC | PYRS | IRI+FU+LEU, IRI+OXA | 100-125mg/m2 (d1, 8, 15 and 22), every 6 weeks; 200mg/m2, every 3 weeks | M | 212 | 61 | 0-2 | 8 |
| Ji41 | 2010 | R | Asian | mCRC | Sequencing | FOLFIRI | 180mg/m2, biweekly | S | 64 | NR | 0-2 | 7 |
| Balibrea31 | 2010 | P | Caucasian | mCRC | Sequencing | IRI+ 5-FU, IRI+5FU/LV | 80mg/m2, weekly; 180mg/m2, biweekly | M | 149 | NR | 0-2 | 8 |
| Han39 | 2009 | P | Asian | mNSCLC | SBE | IRI+CDDP | 65 or 80mg/m2 (d1 and 8), every 3 weeks | S | 107 | 58 | 0-2 | 7 |
| Onoue40 | 2009 | P | Asian | Mixed tumors | Direct Sequencing | IRI alone; IRI+plat; IRI+ other anticancer agents, FOLFIRI | 60-100mg/m2 | S | 133 | NR | 0-1 | 7 |
| Ferraldeschi18 | 2009 | P | Mixed | mCRC | SPR | IRI, FOLFIRI, IRI+VEGF inhibitor | 350mg/m2, every 3 weeks; 180mg/m2, biweekly | S | 92 | 62.9 | NR | 8 |
| Rouits29 | 2008 | R | Caucasian | mCRC | PYRS | FOLFIRI | 180mg/m2, biweekly | S | 44 | 60 | 0-2 | 8 |
| Parodi28 | 2008 | P | Caucasian | mCRC | SPR | FOLFIRI, mIFL, CapeIRI | 125 or 180mg/m2, biweekly; 250mg/m2, every 3 weeks | M | 110 | NR | 0-2 | 8 |
| Liu16 | 2008 | R | Asian | mCRC | SPR | FOLFIRI | 180mg/m2, biweekly | S | 128 | NR | 0-2 | 8 |
| Kweekel15 | 2008 | R | Caucasian | mCRC | PYRS | IRI+CAP+OAX | 250 or 350mg/m2 (d1), every 3 weeks | M | 218 | NR | 0-2 | 8 |
| Wang38 | 2007 | P | Asian | mCRC | SPR | FOLFIRI | 180mg/m2, biweekly | M | 70 | NR | 0-3 | 8 |
| Ruzzo30 | 2007 | P | Caucasian | mCRC | SPR | FOLFIRI | 180mg/m2, biweekly | M | 146 | 61 | NR | 7 |
| Jada37 | 2007 | NR | Asian | Mixed tumors | SPR | IRI | 375 mg/m2, every 3 weeks | S | 45 | 55 | 0-2 | 7 |
| Cote14 | 2007 | P | Caucasian | stage III colon cancer | SPR | LV5FU2+IRI | 180 mg/m2 (d1), every 2 weeks | M | 89 | NR | NR | 8 |
| Toffoli11 | 2006 | P | Caucasian | mCRC | PYRS | mFOLFIRI or FOLFIRI | 180mg/m2 (d1), every 2 weeks | M | 250 | 60.6 | 0-2 | 8 |
| Massacesi12 | 2005 | P | Caucasian | mCRC | Sequencing | IRI+RAL | 80 weekly (d1, 8, 15 and 22), every 5 weeks | M | 56 | 64 | 0-2 | 7 |
| Jong13 | 2006 | P | Caucasian | Mixed tumors | SPR | IR+NEO | 350mg/m2, every 3 weeks | M | 52 | 58 | 0-2 | 8 |
| Han36 | 2006 | P | Asian | mNSCLC | Direct Sequencing | IRI+CDDP | 80mg/m2 (d1 and 8), every 3 weeks | S | 81 | NR | 0-2 | 8 |
| Rouits27 | 2004 | R | Caucasian | mCRC | PYRS | IRIFUFOL, FOLFIRI | 85mg/m2, weekly; 180mg/m2, biweekly | S | 73 | 62 | 0-2 | 8 |
| Marcuello26 | 2004 | P | Caucasian | mCRC | SPR | IRI alone, IRI+TOM, IRI+5-FU, IRI+5-FU+leuc | 80mg/m2, weekly; 180mg/m2, biweekly;3 50mg/m2, every 3 weeks | S | 95 | 68 | 0-2 | 8 |
| Innocenti67 | 2004 | P | Mixed | Mixed tumors | SBE | IRI | 350mg/m2, every 3 weeks | S | 59 | 60 | NR | 7 |
| Font66 | 2003 | NR | NR | mNSCLC | Sequencing | IRI+DOC | 70mg/m2 (d1, 8 and 15), every 4 weeks | S | 47 | 55 | 0-2 | 7 |
| Iyer25 | 2002 | P | Caucasian | Mixed tumors | SPR | IRI | 300mg/m2, every 3 weeks | S | 20 | NR | NR | 8 |
R, analysis was planned retrospectively; P, analysis was planned prospectively; NR, Not reported; mCRC, metastatic colorectal cancer; GC, gastric cancer; SCLC, small-cell lung cancer; NSCLC, non-small-cell lung cancer; SPR, Sizing of PCR products (analysis of fragment size); PYRS, Pyrosequencing; SBE, Single base prime extension assay; IRI, irinotecan; CDDP, cisplatin; BEV, bevacizumab; OXA, oxaliplatin; CET, cetuximab; PLA, platinum; IFL, FU+IRI; CAP, capecitabine; CBP, carboplatin; LOB, lobaplatin; RAL, raltitrexed; 5-FU, 5-fluorouracil; LV, leucovorin; GCB, gemicitabine; TOM, toumdex; DOC, docetaxel; PAC, paclitaxel; IFL, IRI+5-FU/LV; FOLFIRI, FOL stands for folinic acid, F for fluorouracil, IRIR for irinotecan+5-FU; S, single center; M, multicenter; ECOG, Estern Cooperative Oncology Group; NOS: Newcastle-Ottawa Scale.
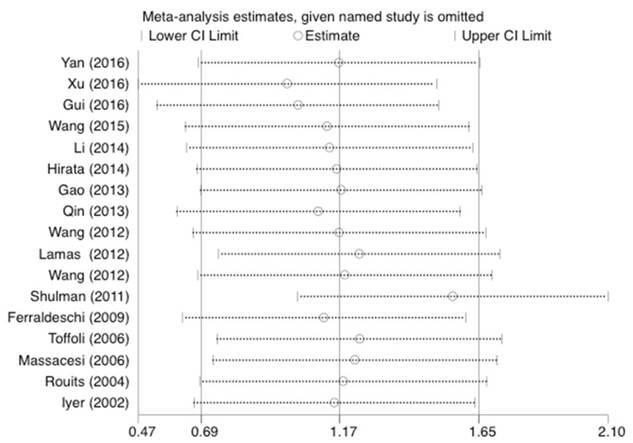
Forest plot of diarrhea risk related to UGT1A1*28 polymorphism under the homozygous model.
Meta-analysis Results for diarrhea.
| Compared genotype | Group | No. of studies | No. of participants | OR (95%CI) | P | Test for heterogeneity | |
|---|---|---|---|---|---|---|---|
| P | I2 | ||||||
| TA6/7 vs. TA6/6 | All | 28 | 3435 | 1.56 (1.25-1.96) | <0.001 | 0.175 | 19.9% |
| mCRC | 16 | 2563 | 1.60 (1.11-2.31) | 0.011 | 0.034 | 43.3% | |
| SCLC | 3 | 131 | 2.40 (0.74-7.74) | 0.144 | 0.208 | 36.3% | |
| mNSCLC | 3 | 235 | 0.92 (0.34-2.54) | 0.879 | 0.883 | 0.0% | |
| Asian | 18 | 2270 | 1.85 (1.37-2.50) | <0.001 | 0.334 | 10.1% | |
| Caucasian | 9 | 1118 | 1.28 (0.91-1.80) | 0.117 | 0.136 | 35.3% | |
| Retrospective | 13 | 2123 | 1.70 (1.09-2.66) | 0.020 | 0.032 | 46.8% | |
| Prospective | 12 | 1090 | 1.69 (1.13-2.52) | 0.010 | 0.495 | 0.0% | |
| TA7/7 vs. TA6/6 | All | 17 | 2610 | 3.97 (1.88-8.38) | <0.001 | 0.007 | 51.7% |
| mCRC | 14 | 1172 | 3.53 (1.54-8.09) | 0.003 | 0.004 | 57.5% | |
| Asian | 10 | 1805 | 8.98 (5.21-15.47) | <0.001 | 0.152 | 32.0% | |
| Caucasian | 7 | 805 | 1.09 (0.56-2.13) | 0.807 | 0.259 | 22.3% | |
| Retrospective | 9 | 1737 | 4.84 (1.32-17.69) | 0.017 | <0.001 | 71.7% | |
| Prospective | 7 | 743 | 2.86 (1.30-6.30) | 0.009 | 0.555 | 0.0% | |
| TA6/7+7/7 vs. TA6/6 | All | 44 | 4868 | 2.18 (1.68-2.83) | <0.001 | 0.003 | 40.8% |
| SCLC | 3 | 131 | 3.95 (1.42-11.01) | 0.009 | 0.115 | 53.8% | |
| mNSCLC | 4 | 321 | 1.24 (0.58-2.65) | 0.582 | 0.560 | 0.0% | |
| Advanced OC | 2 | 178 | 7.09 (2.91-17.26) | <0.001 | 1.00 | 0.0% | |
| mCRC | 25 | 3477 | 1.96 (1.42-2.70) | <0.001 | 0.005 | 47.3% | |
| Asian | 32 | 3607 | 2.74 (2.21-3.40) | <0.001 | 0.132 | 22.2% | |
| Caucasian | 11 | 1214 | 1.39 (0.84-2.32) | 0.202 | 0.038 | 47.9% | |
| Retrospective | 16 | 2359 | 2.17 (1.36-3.49) | 0.001 | 0.001 | 62.0% | |
| Prospective | 24 | 2198 | 2.12 (1.62-2.79) | <0.001 | 0.263 | 14.3% | |
| TA7/7 vs. TA6/7+TA6/6 | All | 24 | 3175 | 3.64 (2.01-6.58) | <0.001 | <0.001 | 57.6% |
| SCLC | 2 | 64 | 19.90 (2.57-154.1) | 0.004 | 0.832 | 0.0% | |
| mCRC | 17 | 2656 | 3.16 (1.61-6.19) | 0.001 | <0.001 | 64.1% | |
| Asian | 13 | 1917 | 8.64 (4.14-18.04) | <0.001 | 0.092 | 36.3% | |
| Caucasian | 10 | 1211 | 1.62 (1.03-2.53) | 0.035 | 0.188 | 27.8% | |
| Retrospective | 11 | 2003 | 2.06 (1.23-3.44) | 0.006 | 0.168 | 32.5% | |
| Prospective | 11 | 995 | 2.92 (1.64-5.21) | <0.001 | 0.219 | 26.2% | |
mCRC, metastatic colorectal cancer; mNSCLC, metastatic non-small-cell lung cancer.
Meta-Analysis of UGT1A1*28 Polymorphism and Response
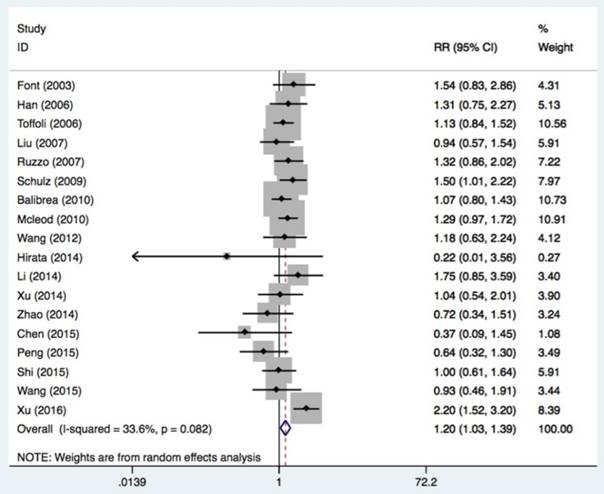
Eighteen studies with 2024 patients were assessed to determine the association of the UGT1A1*28 polymorphism with tumor response to irinotecan-based chemotherapy (Table 4 and Figure 4). A partial or complete remission was grouped as a response, while stable tumor or progression was considered no response. A response occurred in patients with at least one mutation allele but not in patients with the wide genotype (TA6/7+TA7/7 vs. TA6/6: OR = 1.20, 95%CI = 1.07-1.34, P = 0.016). The association was significant in Caucasians (OR = 1.23, 95%CI = 1.06-1.42, P = 0.006), retrospective study designs (OR = 1.54, 95%CI = 1.06-2.23, P = 0.022), and mCRC patients (OR = 1.24, 95%CI = 1.05-1.48, P = 0.014).
Heterogeneity Analysis
There was high heterogeneity among studies evaluating severe diarrhea under the homozygous and recessive comparisons (TA7/7 vs. TA6/6: P = 0.007, I2= 51.7%; TA7/7 vs. TA6/6+TA6/7: P<0.001, I2= 57.6%). We performed meta-regression to explore the sources of heterogeneity. The data indicated that ethnicity and year of publication accounted for 76% and 26% of heterogeneity under the homozygous model and 54% and 41% under the recessive model, respectively (data not shown). There was high heterogeneity among studies of neutropenia under recessive comparison (P<0.001, I2= 60.7%). The meta-regression results only revealed that the number of patients represented 25% of the heterogeneity and no other factors were found (data not shown).
Meta-analysis Results for neutropenia.
| Compared genotype | Group | No. of studies | No. of participants | OR (95%CI) | P | Test for heterogeneity | |
|---|---|---|---|---|---|---|---|
| P | I2 | ||||||
| TA6/7 vs. TA6/6 | All | 32 | 3948 | 1.71 (1.41-2.08) | < 0.001 | 0.104 | 24.8% |
| mCRC | 19 | 2801 | 1.76 (1.40-2.23) | <0.001 | 0.434 | 1.8% | |
| mNSCLC | 2 | 188 | 1.35 (0.55-3.34) | 0.518 | 0.920 | 0.0% | |
| Asian | 21 | 2547 | 1.56 (1.07-2.27) | 0.020 | 0.011 | 46.0% | |
| Caucasian | 10 | 1342 | 1.86 (1.34-2.60) | <0.001 | 0.991 | 0.0% | |
| Retrospective | 14 | 1468 | 1.90 (1.43-2.53) | <0.001 | 0.201 | 23.3% | |
| Prospective | 15 | 1448 | 1.53 (1.15-2.05) | 0.004 | 0.882 | 0.0% | |
| TA7/7 vs. TA6/6 | All | 27 | 3575 | 5.34 (3.05-9.33) | <0.001 | 0.003 | 48.7% |
| mCRC | 19 | 2801 | 5.07 (2.56-10.02) | <0.001 | 0.001 | 59.3% | |
| Asian | 15 | 2154 | 4.77 (1.71-13.22) | 0.003 | 0.001 | 62.6% | |
| Caucasian | 11 | 1362 | 5.39 (3.43-8.47) | <0.001 | 0.342 | 10.7% | |
| Retrospective | 12 | 1914 | 5.61 (3.58-8.82) | <0.001 | <0.001 | 69.3% | |
| Prospective | 14 | 1531 | 5.81 (3.57-9.47) | <0.001 | 0.291 | 14.8% | |
| TA6/7+7/7 vs. TA6/6 | All | 49 | 5232 | 2.15 (1.71-2.70) | <0.001 | 0.003 | 39.5% |
| mCRC | 26 | 3473 | 2.47 (1.86-3.27) | <0.001 | 0.013 | 42.1% | |
| Advanced esophageal cancer | 2 | 133 | 1.20 (0.48-3.05) | 0.697 | 0.691 | 0.0% | |
| Advanced GC | 4 | 193 | 1.40 (0.64-3.06) | 0.402 | 0.759 | 0.0% | |
| mNSCLC | 4 | 351 | 1.79 (0.97-3.33) | 0.064 | 0.432 | 0.0% | |
| Asian | 35 | 3715 | 2.11 (1.54-2.89) | <0.001 | <0.001 | 53.9% | |
| Caucasian | 13 | 1458 | 2.29 (1.69-3.08) | <0.001 | 0.992 | 0.0% | |
| Retrospective | 18 | 2318 | 2.52 (1.64-3.88) | <0.001 | <0.001 | 59.3% | |
| Prospective | 29 | 2739 | 1.90 (1.53-2.35) | <0.001 | 0.530 | 0.0% | |
| TA7/7 vs. TA6/6+6/7 | All | 28 | 3668 | 4.12 (2.36-7.20) | <0.001 | <0.001 | 60.7% |
| mCRC | 20 | 2894 | 3.70 (1.88-7.30) | <0.001 | <0.001 | 69.4% | |
| Asian | 15 | 2154 | 4.16 (1.44-11.99) | 0.008 | <0.001 | 68.9% | |
| Caucasian | 12 | 1455 | 3.39 (1.92-5.98) | <0.001 | 0.057 | 42.7% | |
| Retrospective | 12 | 1914 | 3.59 (1.05-12.28) | 0.042 | <0.001 | 76.4% | |
| Prospective | 15 | 1624 | 4.10 (2.36-7.12) | <0.001 | 0.088 | 35.1% | |
mCRC, metastatic colorectal cancer; GC, gastric cancer; mNSCLC, metastatic non-small-cell lung cancer.
Meta-analysis Results for response.
| Group | No. of studies | No. of participants | OR (95%CI) | P | Test for heterogeneity | |
|---|---|---|---|---|---|---|
| P | I2 | |||||
| All | 18 | 2024 | 1.20 (1.07-1.34) | 0.016 | 0.082 | 33.6% |
| mCRC | 12 | 1691 | 1.24 (1.05-1.48) | 0.014 | 0.060 | 42.2% |
| SCLC | 2 | 64 | 0.87 (0.57-1.33) | 0.514 | 0.458 | 0.0% |
| mNSCLC | 3 | 202 | 1.08 (0.71-1.63) | 0.726 | 0.127 | 51.5% |
| Asian | 12 | 2270 | 1.08 (0.82-1.42) | 0.168 | 0.019 | 51.7% |
| Caucasian | 5 | 1118 | 1.23 (1.06-1.42) | 0.006 | 0.669 | 0.0% |
| Retrospective | 4 | 538 | 1.54 (1.06-2.23) | 0.022 | 0.060 | 59.5% |
| Prospective | 12 | 1292 | 1.07 (0.93-1.22) | 0.343 | 0.511 | 0.0% |
mCRC, metastatic colorectal cancer; mNSCLC, metastatic non-small-cell lung cancer
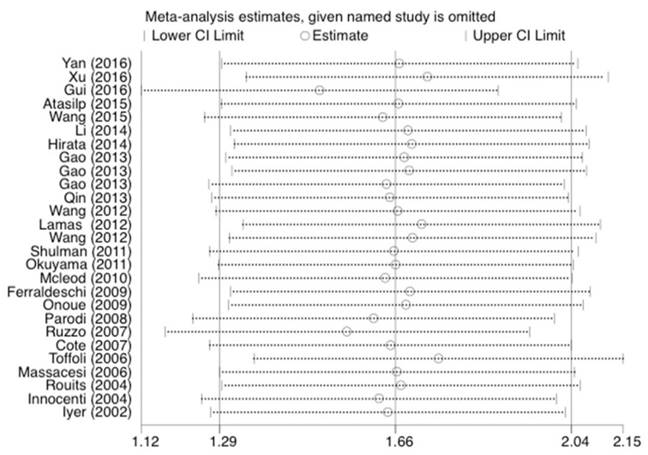
Forest plot of neutropenia risk related to UGT1A1*28 polymorphism under the homozygous model.
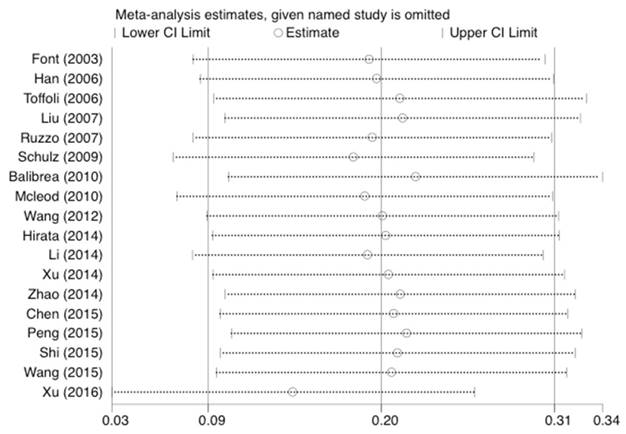
Forest plot of response related to UGT1A1*28 polymorphism under the homozygous model.
P values for Begg's funnel plot and Egger's test for diarrhea and neutropenia.
| Begg | Egger | |
|---|---|---|
| Diarrhea | ||
| TA6/7 vs. TA6/6 | 0.635 | 0.244 |
| TA7/7 vs. TA6/6 | 0.365 | 0.166 |
| TA6/7+TA7/7 vs. TA6/6 | 0.927 | 0.282 |
| TA7/7 vs. TA6/6+TA6/7 | 0.215 | 0.697 |
| Neutropenia | ||
| TA6/7 vs. TA6/6 | 0.284 | 0.088 |
| TA7/7 vs. TA6/6 | 0.755 | 0.999 |
| TA6/7+TA7/7 vs. TA6/6 | 0.044 | 0.027 |
| TA7/7 vs. TA6/6+TA6/7 | 0.782 | 0.617 |
Publication Bias
To detect publication bias in studies that evaluated diarrhea and neutropenia, we performed the Begg and Egger tests (Table 5). As shown in Table 5, publication bias was found only among the studies of neutropenia under the dominant model (P = 0.027). Next, a trim and fill method was applied and the results (OR = 1.80, 95%CI = 1.37-2.36, P<0.001) showed no statistical difference compared from the results described above (OR = 2.15, 95%CI = 1.71-2.70, P<0.001). There was also no publication bias in studies evaluating response (P = 0.082). Thus, publication bias did not appear to affect our results.
Sensitivity Analysis
Statistical analysis was conducted as described previously [23]. As shown in Figure 5, 6, and 7, the results were not affected by omitting individual studies in this meta-analysis, indicating that our results are reliable.
Trial Sequential analysis
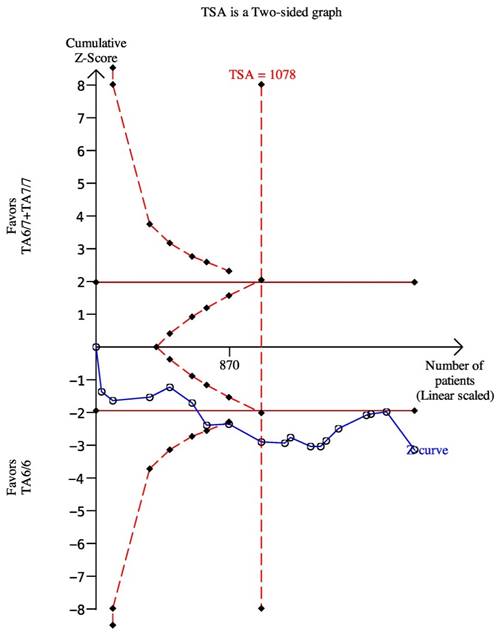
We used the dominant model as an example to perform the TSA, which included eighteen trials with 2024 patients. The results showed the required information size was 1078, which meant our sample size was enough to get a robust conclusion about the UGT1A1*28 polymorphism and chemotherapy response (Figure 8). The required sample sizes for determining the associations between UGT1A1 and diarrhea and neutropenia under the dominant model were 763 and 1162, respectively (data were not shown).
Sensitivity analysis of the studies about diarrhea under the homozygous model.
Sensitivity analysis of the studies about neutropenia under the homozygous model.
Sensitivity analysis of the studies about response under the dominant model.
The required sample size to demonstrate the relationship between UGT11A1*28 polymorphism and chemotherapy response. The solid line represents the cumulative z-curve. The dashed curve represents the trial sequential monitoring boundary.
Discussion
A couple of meta-analyses have investigated the relationships between the UGT1A1*28 polymorphism and irinotecan-induced toxicity, severe diarrhea, and neutropenia. A study by Chen et al. in 2014 included six articles and found no statistically significant association between the UGT1A1*28 polymorphism and neutropenia in Asians (OR = 1.67, 95%CI = 0.94-2.97) [65]. Liu et al.[66] conducted a meta-analysis of 16 articles and found that mCRC patients carrying the TA7/7 genotype had a higher risk of neutropenia and diarrhea in Caucasians. In contrast to previous studies, we evaluated 58 articles including 6087 cancer patients and performed stratified analyses based on ethnicity, study design, and cancer type. Statistical difference between the UGT1A1*28 polymorphism and diarrhea was confirmed in Asian patients and mCRC patients under the five models. Individuals with at least mutation allele had a 1.71- and 5.34-fold greater risk of neutropenia than individuals carrying the wide genotype. Mutated genotypes of the UGT1A1*28 polymorphism may lower the glucuronidation rates of SN-38 and lead to greater susceptibility to severe toxicities [25, 36].
Patients evaluated in this study, particularly mCRC patients with the TA6/7 and TA7/7 genotypes, may have severe diarrhea and neutropenia after irinotecan-induced chemotherapy. However, the UGT1A1*28 TA6/6 and TA7/7 genotypes may show an increased treatment response according to our results. In contrast to our results, Xu et al.[67] observed different clinical responses in Ugyur patients with different UGT1A1*28 polymorphism genotypes, but not in the Han population. Although the reduction of irinotecan was greater in patients with the TA7/7 or TA6/7 genotypes than the TA6/6 genotype, no difference in overall or progression-free survival between the two group patients were found by Dias et al.[68]. These results indicate that if the patients with mutant genotypes could tolerate the toxicities, irinotecan-based chemotherapy is a good choice for treatment. Additional studies of the treatment response should be carried out.
Previous meta-analyses included few than 20 studies and only focused on toxicities or chemotherapy response. In comparison with these studies, we included more research (58 studies) and investigated the associations of UGT1A1*28 polymorphism with toxicities and chemotherapy effect. We also got a novel conclusion that patients with a higher risk of chemotherapy toxicities have a tendency to better response to chemotherapy. However, there were some limitations to our study. First, the number of studies of SCLC, mNSCLC, advanced GC, solid tumors, and other cancers were limited, and thus, larger sample sizes for a single tumor are needed to validate our results. Second, high heterogeneity existed among studies related to severe neutropenia under the recessive comparison. Although the number of patients could explain 25% of the heterogeneity, other influencing factors were not identified. Third, the studies we including selected different irinotecan doses in the chemotherapies, which may lead to some bias.
Conclusions
In conclusion, we detected a significant relationship between the UGT1A1*28 polymorphism and irinotecan-induced toxicity and response to irinotecan-based chemotherapy. This polymorphism may be useful as a detective index for cancer patients receiving irinotecan-based chemotherapy.
Acknowledgements
This study was supported by National Natural Science Foundation, China (No. 81471670; 81274136); China Postdoctoral Science Foundation (No. 2014M560791; 2015T81037); Science and Technology Plan of Innovation Project, Shaanxi Province, People's Republic of China (No 2015KTCL03-06) and the Fundamental Research Funds for the Central Universities, China (No. 2014qngz-04).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA: a cancer journal for clinicians. 2016;66:7-30
2. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. et al. Cancer statistics in China, 2015. CA: a cancer journal for clinicians. 2016;66:115-32
3. Xu Q, Ding YY, Song LX, Xu JF. Correlation of UGT1A1 and ERCC1 gene polymorphisms with the outcome of combined irinotecan plus cisplatin treatment in recurrent ovarian cancer. Genetics and molecular research: GMR. 2015;14:7241-7
4. Li J, Yu Q, Fu S, Xu M, Zhang T, Xie C. et al. A novel genetic score model of UGT1A1 and TGFB pathway as predictor of severe irinotecan-related diarrhea in metastatic colorectal cancer patients. Journal of cancer research and clinical oncology. 2016;142:1621-8
5. Wang W, Huang J, Tao Y, Lyu X, Yang L, Wu D. et al. Phase II and UGT1A1 Polymorphism Study of Two Different Irinotecan Dosages Combined with Cisplatin as First-Line Therapy for Advanced Gastric Cancer. Chemotherapy. 2016;61:197-203
6. Fujita K, Sparreboom A. Pharmacogenetics of irinotecan disposition and toxicity: a review. Current clinical pharmacology. 2010;5:209-17
7. Gupta E, Lestingi TM, Mick R, Ramirez J, Vokes EE, Ratain MJ. Metabolic fate of irinotecan in humans: correlation of glucuronidation with diarrhea. Cancer research. 1994;54:3723-5
8. Yan L, Wang XF, Wei LM, Nie YL, Liu JY, Zhang LR. Effects of UGT1A1*6, UGT1A1*28, and ABCB1-3435C>T polymorphisms on irinotecan induced toxicity in Chinese cancer patients. International journal of clinical pharmacology and therapeutics. 2016;54:193-9
9. Xiao XG, Xia S, Zou M, Mei Q, Zhou L, Wang SJ. et al. The relationship between UGT1A1 gene polymorphism and irinotecan effect on extensive-stage small-cell lung cancer. OncoTargets and therapy. 2015;8:3575-83
10. Atasilp C, Chansriwong P, Sirachainan E, Reungwetwattana T, Chamnanphon M, Puangpetch A. et al. Correlation of UGT1A1(*)28 and (*)6 polymorphisms with irinotecan-induced neutropenia in Thai colorectal cancer patients. Drug metabolism and pharmacokinetics. 2016;31:90-4
11. Toffoli G, Cecchin E, Corona G, Russo A, Buonadonna A, D'Andrea M. et al. The role of UGT1A1*28 polymorphism in the pharmacodynamics and pharmacokinetics of irinotecan in patients with metastatic colorectal cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2006;24:3061-8
12. Massacesi C, Terrazzino S, Marcucci F, Rocchi MB, Lippe P, Bisonni R. et al. Uridine diphosphate glucuronosyl transferase 1A1 promoter polymorphism predicts the risk of gastrointestinal toxicity and fatigue induced by irinotecan-based chemotherapy. Cancer. 2006;106:1007-16
13. de Jong FA, Kehrer DF, Mathijssen RH, Creemers GJ, de Bruijn P, van Schaik RH. et al. Prophylaxis of irinotecan-induced diarrhea with neomycin and potential role for UGT1A1*28 genotype screening: a double-blind, randomized, placebo-controlled study. The oncologist. 2006;11:944-54
14. Cote JF, Kirzin S, Kramar A, Mosnier JF, Diebold MD, Soubeyran I. et al. UGT1A1 polymorphism can predict hematologic toxicity in patients treated with irinotecan. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13:3269-75
15. Kweekel DM, Gelderblom H, Van der Straaten T, Antonini NF, Punt CJ, Guchelaar HJ. UGT1A1*28 genotype and irinotecan dosage in patients with metastatic colorectal cancer: a Dutch Colorectal Cancer Group study. British journal of cancer. 2008;99:275-82
16. Liu CY, Chen PM, Chiou TJ, Liu JH, Lin JK, Lin TC. et al. UGT1A1*28 polymorphism predicts irinotecan-induced severe toxicities without affecting treatment outcome and survival in patients with metastatic colorectal carcinoma. Cancer. 2008;112:1932-40
17. Hirata K, Nagata N, Kato T, Okuyama Y, Andoh H, Takahashi K. et al. Prospective phase II trial of second-line FOLFIRI in patients with advanced colorectal cancer including analysis of UGT1A1 polymorphisms: FLIGHT 2 study. Anticancer research. 2014;34:195-201
18. Ferraldeschi R, Minchell LJ, Roberts SA, Tobi S, Hadfield KD, Blackhall FH. et al. UGT1A1*28 genotype predicts gastrointestinal toxicity in patients treated with intermediate-dose irinotecan. Pharmacogenomics. 2009;10:733-9
19. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Journal of clinical epidemiology. 2009;62:1006-12
20. Yang XL, Xie S, Jiang YY, Shi C, Cai ZG, Chen SX. Association between CYP1A1 Ile462Val Polymorphism and Oral Squamous Cell Carcinoma Susceptibility: Evidence from 13 Investigations. Journal of Cancer. 2015;6:302-9
21. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. Bmj. 1997;315:629-34
22. Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088-101
23. Liu XH, Dai ZM, Kang HF, Lin S, Ma XB, Wang M. et al. Association of IL-23R Polymorphisms (rs6682925, rs10889677, rs1884444) With Cancer Risk: A PRISMA-Compliant Meta-Analysis. Medicine. 2015;94:e2361
24. Duval S, Tweedie R. Trim and fill: A simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56:455-63
25. Iyer L, Das S, Janisch L, Wen M, Ramirez J, Karrison T. et al. UGT1A1*28 polymorphism as a determinant of irinotecan disposition and toxicity. The pharmacogenomics journal. 2002;2:43-7
26. Marcuello E, Altes A, Menoyo A, Del Rio E, Gomez-Pardo M, Baiget M. UGT1A1 gene variations and irinotecan treatment in patients with metastatic colorectal cancer. British journal of cancer. 2004;91:678-82
27. Rouits E, Boisdron-Celle M, Dumont A, Guerin O, Morel A, Gamelin E. Relevance of different UGT1A1 polymorphisms in irinotecan-induced toxicity: a molecular and clinical study of 75 patients. Clinical cancer research: an official journal of the American Association for Cancer Research. 2004;10:5151-9
28. Parodi L, Pickering E, Cisar LA, Lee D, Soufi-Mahjoubi R. Utility of Pretreatment Bilirubin Level and UGT1A1 Polymorphisms in Multivariate Predictive Models of Neutropenia Associated with Irinotecan Treatment in Previously Untreated Patients with Colorectal Cancer. Archives of drug information. 2008;1:97-106
29. Rouits E, Charasson V, Petain A, Boisdron-Celle M, Delord JP, Fonck M. et al. Pharmacokinetic and pharmacogenetic determinants of the activity and toxicity of irinotecan in metastatic colorectal cancer patients. British journal of cancer. 2008;99:1239-45
30. Ruzzo A, Graziano F, Loupakis F, Santini D, Catalano V, Bisonni R. et al. Pharmacogenetic profiling in patients with advanced colorectal cancer treated with first-line FOLFIRI chemotherapy. The pharmacogenomics journal. 2008;8:278-88
31. Martinez-Balibrea E, Abad A, Martinez-Cardus A, Gines A, Valladares M, Navarro M. et al. UGT1A and TYMS genetic variants predict toxicity and response of colorectal cancer patients treated with first-line irinotecan and fluorouracil combination therapy. British journal of cancer. 2010;103:581-9
32. McLeod HL, Sargent DJ, Marsh S, Green EM, King CR, Fuchs CS. et al. Pharmacogenetic predictors of adverse events and response to chemotherapy in metastatic colorectal cancer: results from North American Gastrointestinal Intergroup Trial N9741. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28:3227-33
33. Shulman K, Cohen I, Barnett-Griness O, Kuten A, Gruber SB, Lejbkowicz F. et al. Clinical implications of UGT1A1*28 genotype testing in colorectal cancer patients. Cancer. 2011;117:3156-62
34. Lamas MJ, Duran G, Balboa E, Bernardez B, Candamio S, Vidal Y. et al. The value of genetic polymorphisms to predict toxicity in metastatic colorectal patients with irinotecan-based regimens. Cancer chemotherapy and pharmacology. 2012;69:1591-9
35. Wang W, Chen Y, Feng F, Zhao L, Hu B, Lin X. et al. The association between UGT1A1*28 gene polymorphism and irinotecan-based chemotherapy in metastatic colorectal cancer. 2015 clinical acute and severe experience exchange Second Summit Forum. 2015:1-1
36. Han JY, Lim HS, Shin ES, Yoo YK, Park YH, Lee JE. et al. Comprehensive analysis of UGT1A polymorphisms predictive for pharmacokinetics and treatment outcome in patients with non-small-cell lung cancer treated with irinotecan and cisplatin. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2006;24:2237-44
37. Jada SR, Lim R, Wong CI, Shu X, Lee SC, Zhou Q. et al. Role of UGT1A1*6, UGT1A1*28 and ABCG2 c.421C>A polymorphisms in irinotecan-induced neutropenia in Asian cancer patients. Cancer science. 2007;98:1461-7
38. Wang Y, Xu J, Shen L, Xu N, Wang J, J S. et al. Polymorphisms of UGT1A gene and irinotecan toxicity in chinese colorectal cancer patients. Clin J Oncol. 2008;29:913-6
39. Han JY, Lim HS, Park YH, Lee SY, Lee JS. Integrated pharmacogenetic prediction of irinotecan pharmacokinetics and toxicity in patients with advanced non-small cell lung cancer. Lung cancer (Amsterdam, Netherlands). 2009;63:115-20
40. Onoue M, Terada T, Kobayashi M, Katsura T, Matsumoto S, Yanagihara K. et al. UGT1A1*6 polymorphism is most predictive of severe neutropenia induced by irinotecan in Japanese cancer patients. International journal of clinical oncology. 2009;14:136-42
41. Ji C, He Y, Hu B, Wang G, Yao Y, Chen J. et al. The association between UGT1A1*28 polymorphism with the efficacy of irinotecan chemotherapy on advanced colorectal cancer and its adverse reaction. Oncology. 2010;30:870-4
42. Nakamura Y, Soda H, Oka M, Kinoshita A, Fukuda M, Fukuda M. et al. Randomized phase II trial of irinotecan with paclitaxel or gemcitabine for non-small cell lung cancer: association of UGT1A1*6 and UGT1A1*27 with severe neutropenia. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2011;6:121-7
43. Okuyama Y, Hazama S, Nozawa H, Kobayashi M, Takahashi K, Fujikawa K. et al. Prospective phase II study of FOLFIRI for mCRC in Japan, including the analysis of UGT1A1 28/6 polymorphisms. Japanese journal of clinical oncology. 2011;41:477-82
44. Park SR, Kong SY, Rhee J, Park YI, Ryu KW, Lee JH. et al. Phase II study of a triplet regimen of S-1 combined with irinotecan and oxaliplatin in patients with metastatic gastric cancer: clinical and pharmacogenetic results. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 2011;22:890-6
45. Wang Y, Shen L, Xu N, Wang JW, Jiao SC, Liu ZY. et al. UGT1A1 predicts outcome in colorectal cancer treated with irinotecan and fluorouracil. World journal of gastroenterology: WJG. 2012;18:6635-44
46. Zhang X, Wang C, Huang M, Fu X, Lu B, Deng Y. et al. Relationship between UGT1A1 gene polymorphisms and toxicity/efficacy of irinotecan-based chemotherapy in metastatic colorectal cancer. Chinese Journal of Pathophysiology. 2012;05:823-8
47. Wang Y, Ge F, Lin L, Hao G, Shen L, Xu N. et al. Correlation between UGT1A1 gene polymorphisms and toxicity and efficacy in patients with metastatic colorectal cancer treated with irinotecan based chemotherapy. Chinese Clinical Oncology. 2012;11:961-6
48. Gao J, Zhou J, Li Y, Lu M, Jia R, Shen L. UGT1A1 6/28 polymorphisms could predict irinotecan-induced severe neutropenia not diarrhea in Chinese colorectal cancer patients. Medical oncology (Northwood, London, England). 2013;30:604
49. Gao J, Zhou J, Li Y, Peng Z, Li Y, Wang X. et al. Associations between UGT1A1*6/*28 polymorphisms and irinotecan-induced severe toxicity in Chinese gastric or esophageal cancer patients. Medical oncology (Northwood, London, England). 2013;30:630
50. Hirasawa A, Zama T, Akahane T, Nomura H, Kataoka F, Saito K. et al. Polymorphisms in the UGT1A1 gene predict adverse effects of irinotecan in the treatment of gynecologic cancer in Japanese patients. Journal of human genetics. 2013;58:794-8
51. Qin Q, Yang L, Zhou A, Huang J, Zhang W, Sun Y. et al. Study of irinotecan-induced toxicity and its correlation to UGT1A1*28 gene promoter polymorphisms. Oncology progress. 2013;04:369-73
52. Zhou CF, Ma T, Su Y, Ye ZB, Ji J, Yu YY. et al. UGT1A1 gene polymorphisms and the toxicities of FOLFIRI in Chinese Han patients with gastrointestinal cancer. Anti-cancer agents in medicinal chemistry. 2013;13:235-41
53. Xu H, Zhu X, Wang L, Zhu C. Correlation of UGT1A1 gene polymorphism with the efficacy and toxicity of Irinotecan for Chemotherapy of Gastric cancer. Journal of Clinical Research. 2014;01:72-4
54. Li M, Wang Z, Guo J, Liu J, Li C, Liu L. et al. Clinical significance of UGT1A1 gene polymorphisms on irinotecan-based regimens as the treatment in metastatic colorectal cancer. OncoTargets and therapy. 2014;7:1653-61
55. Zhao X, Song M, Xu J, Wang Y. Correlation between UGT1A1 gene polymorphisms and toxicity and efficacy of irinotecan chemotherapy on small cell lung cancer patients. Journal of Hebei Medical University. 2014;09:1000-2
56. Song X, Xu Q, Ding Y, Xu J, Gao J, Ma J. The relationship of UGT1A1 and ERCC1 gene polymorphism with Irinotecan and Cisplatin Combination Regimen for Advanced Ovary Cancer. Beijing Medical Journal. 2014;07:530-3
57. Zhang Y, Su D, Zhang T, Mao Z, Bai L. The relationship between UGT1A1 gene polymorphisms and the toxicity and efficacy of irinotecan-based chemotherapy in advanced colorectal cancer. Chinese Journal of Drug Application and Monitoring. 2014;05:263-6
58. Zhou Y, Zhuang R, Chen P, Tan Q, Zhang C, Guo W. et al. Relationship between UGT1A1 gene polymorphisms and toxicity of irinotecan-based chemotherapy in colorectal cancer. CHINA ONCOLOGY. 2014;07:493-500
59. Wu H, Zhou D, Li J. Correlation of UGT1A1 polymorphism and safety of Irinotecan-based regimens treatment in Advanced Esophageal Cancer. Strait Pharmaceutical Journal. 2015;01:60-2
60. Peng J, Deng Y, Ma D, Liao W, He Y. The influence of UGT1A*28 gene polymorphism on efficacy and adverse effect of combined irinotecan chemotherapy for late-staged colorectal cancer, a preliminary report in southern China. J Dig Oncol. 2015:28-32
61. Shi Y, Hu Y, Hu X, Li X, Lin L, Han X. Cisplatin combined with irinotecan or etoposide for untreated extensive-stage small cell lung cancer: A multicenter randomized controlled clinical trial. Thoracic cancer. 2015;6:785-91
62. Chen Y, Zhang W, Ji Z. Toxicity and efficacy relationship between UGT1A1 gene polymorphisms and irinotecan-based chemotherapy in non-small cell lung cancer. Journal of Qiqihar University of Medicine. 2015;22:3277-80
63. Font A, Sanchez JM, Taron M, Martinez-Balibrea E, Sanchez JJ, Manzano JL. et al. Weekly regimen of irinotecan/docetaxel in previously treated non-small cell lung cancer patients and correlation with uridine diphosphate glucuronosyltransferase 1A1 (UGT1A1) polymorphism. Investigational new drugs. 2003;21:435-43
64. Innocenti F, Undevia SD, Iyer L, Chen PX, Das S, Kocherginsky M. et al. Genetic variants in the UDP-glucuronosyltransferase 1A1 gene predict the risk of severe neutropenia of irinotecan. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2004;22:1382-8
65. Chen YJ, Hu F, Li CY, Fang JM, Chu L, Zhang X. et al. The association of UGT1A1*6 and UGT1A1*28 with irinotecan-induced neutropenia in Asians: a meta-analysis. Biomarkers: biochemical indicators of exposure, response, and susceptibility to chemicals. 2014;19:56-62
66. Liu X, Cheng D, Kuang Q, Liu G, Xu W. Association of UGT1A1*28 polymorphisms with irinotecan-induced toxicities in colorectal cancer: a meta-analysis in Caucasians. The pharmacogenomics journal. 2014;14:120-9
67. Xu C, Tang X, Qu Y, Keyoumu S, Zhou N, Tang Y. UGT1A1 gene polymorphism is associated with toxicity and clinical efficacy of irinotecan-based chemotherapy in patients with advanced colorectal cancer. Cancer chemotherapy and pharmacology. 2016;78:119-30
68. Dias MM, Pignon JP, Karapetis CS, Boige V, Glimelius B, Kweekel DM. et al. The effect of the UGT1A1*28 allele on survival after irinotecan-based chemotherapy: a collaborative meta-analysis. The pharmacogenomics journal. 2014;14:424-31
Author contact
![]() Corresponding author: Zhi-Jun Dai, Department of Oncology, Second Affiliated Hospital of Xi'an Jiaotong University, Xi'an 710004, China (E-Mail: dzj0911com).
Corresponding author: Zhi-Jun Dai, Department of Oncology, Second Affiliated Hospital of Xi'an Jiaotong University, Xi'an 710004, China (E-Mail: dzj0911com).

 Global reach, higher impact
Global reach, higher impact