3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(2):266-277. doi:10.7150/jca.17379 This issue Cite
Research Paper
Association of MTRR A66G polymorphism with cancer susceptibility: Evidence from 85 studies
1. The Key Laboratory of Pharmacology and Medical Molecular Biology, Medical College, Henan University of Science and Technology, Luoyang 471023, Henan, China;
2. The Molecular Medicine Key Laboratory of Liver Injury and Repair, Medical College, Henan University of Science and Technology, Luoyang 471023, Henan, China;
3. Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, Guangzhou 510623, Guangdong, China.
* These authors contributed equally to this work.
Received 2016-8-29; Accepted 2016-11-14; Published 2017-1-15
Abstract
Methionine synthase reductase (MTRR) is a key regulatory enzyme involved in the folate metabolic pathway. Previous studies investigating the association of MTRR A66G polymorphism with cancer susceptibility reported inconclusive results. We performed the current meta-analysis to obtain a more precise estimation of the possible association. Published literatures were identified from PubMed, Embase and CBM databases up to October 2016. The strength of the association between the MTRR A66G polymorphism and cancer susceptibility was assessed using odds ratios (ORs) and the corresponding 95% confidence intervals (CIs). Eighty five published studies with 32,272 cases and 37,427 controls were included in this meta-analysis. Pooled results indicated that the MTRR A66G polymorphism was associated with an increased overall cancer risk (homozygous model: OR = 1.08, 95% CI = 1.02-1.15, P = 0.009; recessive model: OR = 1.06, 95% CI = 1.00-1.12, P < 0.001 and allele comparison: OR = 1.03, 95% CI = 1.00-1.06, P < 0.001). Stratification analysis further indicated significant associations in head and neck cancer, Caucasians, Africans, and high quality studies. However, to avoid the “false-positive report”, the significant findings were assessed by the false-positive report probability (FPRP) test. Interestingly, the results of FPRP test revealed that the increased risk for MTRR A66G polymorphism among Africans need further validation due to the high probabilities of false-positive results. This meta-analysis suggests that the MTRR A66G polymorphism is associated with significantly increased cancer risk, a finding that needs to be confirmed in single large studies.
Keywords: Methionine synthase reductase (MTRR), polymorphism, susceptibility, meta-analysis.
Introduction
Cancer remains the leading cause of death worldwide, with approximately 14.1 million new cancer cases and 8.2 million deaths occurring in 2012 according to the GLOBOCAN estimates [1]. It has been estimated that about one-third of cancers are attributable to diet and lifestyle [2], and a number of studies have reported a relationship between folate intake and cancer risk [3-5].
Folate plays an important role in one-carbon metabolism, and acts as a coenzyme in DNA methylation and synthesis [6]. Folate can provide the methyl group donor S-adenosylmethionine for many biological reactions. It also plays a critical role in the de novo synthesis of purines and thymidylate, which are necessary for DNA replication and repair [7]. Abnormal folate metabolism can lead to the aberrant distribution of methyl groups and affect DNA biosynthesis and methylation, which is considered as a mechanism in the development of cancer [8].
Methionine synthase reductase (MTRR) is one of the key regulatory enzymes involved in the folate metabolic pathway. It can catalyze the regeneration of methyl cobalamin, which is a cofactor of methionine synthase (MTR) in the remethylation of homocysteine to methionine [9]. Because MTRR plays a vital role in maintaining the active state of MTR, genetic variation within the MTRR gene may be associated with cancer susceptibility. The MTRR gene is located on chromosome 5 at 5p15.2-p15.3, and the most common polymorphism is the substitution of isoleucine with methionine at position 22 (A66G; rs1801394). It has been suggested that the 66GG genotype is negatively correlated with plasma homocysteine levels [10]. A large number of studies have investigated the role of the MTRR A66G polymorphism and cancer risk [11-82], but the results remain controversial. Therefore, we conducted this updated meta-analysis from all eligible studies to derive a more precise estimation of this association.
Materials and methods
Search strategy
A comprehensive literature search was carried out in PubMed, Embase, and Chinese Biomedical (CBM) databases for all relevant articles using the following search terms: “MTRR or methionine synthase reductase or one-carbon metabolism”, “polymorphism or variant or variation” and “cancer or tumor or carcinoma or neoplasm” (the last search was updated on October 21, 2016). Review articles and references cited in the searched studies were examined manually to identify additional relevant articles. Only the most recent study or the one with most participants was included in the final meta-analysis if two or more studies overlapped.
Inclusion and exclusion criteria
The included studies met the following criteria: (1) case-control study design; (2) investigating the association between the MTRR A66G polymorphism and cancer risk; (3) providing detail information for calculating pooled odds ratios (ORs) and their 95% confidence intervals (CIs). Studies were excluded if one of the following existed: (1) not a case-control study; (2) duplicate publications; (3) without detail genotype frequencies; and (4) genotype frequencies in the controls departed from Hardy-Weinberg equilibrium (HWE).
Data extraction
Information was extracted from all eligible studies independently by two authors (Ping Wang and Meilin Wang) according to the inclusion and exclusion criteria listed above. Disagreement was resolved by discussion until consensus was reached. The following information was collected from each study: first author's surname, year of publication, country of origin, ethnicity, cancer type, control source (hospital-based or population-based), genotyping methods, and numbers of cases and controls with the AA, AG and GG genotypes. Ethnicities were categorized as Asians, Caucasians, Africans or Mixed, which included individuals belonging to more than one ethnic group.
Quality assessment
Quality assessment was performed by two authors independently according to the criteria as described previously [83]. Quality scores of studies ranged from 0 (lowest) to 15 (highest), and the studies were categorized into high quality (scores > 9) and low quality (scores ≤ 9).
Statistical analysis
The strength of association between the MTRR A66G polymorphism and cancer risk was assessed by calculating the ORs with the corresponding 95% CIs. The pooled ORs of 5 comparison models were calculated: homozygous model (GG vs. AA), heterozygous model (AG vs. AA), recessive model [GG vs. (AA + AG)], dominant model [(GG +AG) vs. AA] as well as an allele comparison (G vs. A). The Chi square-based Q-test was used to check heterogeneity between studies. A P value greater than 0.1 for the Q-test indicated the homogeneity among studies, in which case the fixed-effects model (the Mantel-Haenszel method) [84] was adopted. Otherwise, the random-effects model (the DerSimonian and Laird method) [85] was applied. Data were stratified by cancer type (if one cancer type was represented by fewer than two studies, it was merged into the “other cancers” group), ethnicity (Asians, Caucasians, Africans or Mixed), source of control (hospital-based studies and population-based studies), and quality scores (≤ 9 and > 9). Potential publication bias was estimated using Begg's funnel plot [86] and Egger's linear regression test [87]. Sensitivity analysis was carried out to evaluate the effect of each individual study on the pooled ORs by excluding studies one-by-one and recalculating the ORs and 95% CIs.
For significant results found in the present meta-analysis, the false-positive report probability (FPRP) was used to evaluate positive associations. We calculated FPRP with 0.2 as a threshold and assigned a prior probability of 0.1 to detect an OR of 0.67/1.50 (protective/risk effects) for an association with genotypes under investigation. FPRP values < 0.2 were considered as noteworthy associations [88]. All the statistical tests were performed with STATA version 12.0 (Stata Corporation, College Station, TX). All the P values were two-sided, and P < 0.05 was considered statistically significant.
Results
Study characteristics
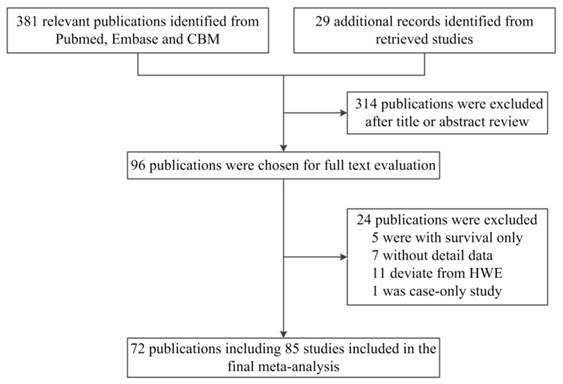
As shown in Figure 1, a total of 381 published records were identified from PubMed, Embase and CBM by using the search terms described above. By checking the reference lists, we identified 29 additional publications. After screening the abstracts and texts, only 96 publications met the crude inclusion criteria and were selected for further assessment. Among them, five were excluded for containing survival data only [89-93], seven lacked detailed data for further analysis [94-100], eleven deviated from HWE [101-111] and one was a case-only study [112]. Ultimately, 72 publications [11-82] were included in the final meta-analysis (Table 1).
Flow diagram of the study selection process.
Characteristics of studies included in the meta-analysis.
| Surname [ref] | Year | Country | Ethnicity | Cancer type | Control | Genotype method | Case | Control | MAF | HWE | Score | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| source | AA | AG | GG | AA | AG | GG | |||||||||
| Le Marchand [11] | 2002 | USA | Asian | Colorectal | PB | PCR-RFLP | 148 | 140 | 26 | 193 | 170 | 30 | 0.29 | 0.374 | 11 |
| Le Marchand [11] | 2002 | USA | Caucasian | Colorectal | PB | PCR-RFLP | 26 | 81 | 40 | 45 | 86 | 39 | 0.48 | 0.865 | 10 |
| Le Marchand [11] | 2002 | USA | Mixed | Colorectal | PB | PCR-RFLP | 30 | 34 | 12 | 40 | 38 | 9 | 0.32 | 0.995 | 9 |
| Stolzenberg-Solomon [12] | 2003 | China | Asian | Esophagus | PB | Real-time PCR | 50 | 63 | 16 | 186 | 179 | 33 | 0.31 | 0.268 | 14 |
| Stolzenberg-Solomon [12] | 2003 | China | Asian | Gastric | PB | Real-time PCR | 43 | 37 | 10 | 186 | 179 | 33 | 0.31 | 0.268 | 13 |
| Gemmati [13] | 2004 | Italy | Caucasian | ALL | PB | PCR-RFLP | 28 | 58 | 23 | 59 | 122 | 76 | 0.47 | 0.457 | 10 |
| Gemmati [13] | 2004 | Italy | Caucasian | NHL | PB | PCR-RFLP | 51 | 106 | 43 | 59 | 122 | 76 | 0.47 | 0.457 | 10 |
| Otani [14] | 2005 | Japan | Asian | Colorectal | HB | Taqman | 58 | 44 | 5 | 128 | 82 | 14 | 0.25 | 0.858 | 8 |
| Shi [15] | 2005 | USA | Caucasian | Lung | HB | PCR-RFLP | 162 | 503 | 370 | 231 | 542 | 375 | 0.44 | 0.168 | 11 |
| Zhang [16] | 2005 | USA | Caucasian | Head and neck | HB | PCR-RFLP | 114 | 376 | 231 | 276 | 589 | 369 | 0.46 | 0.161 | 11 |
| Chen [17] a | 2006 | China | Asian | Colorectal | PB | PCR-RFLP | 32 | 107 (AG+GG) | 89 | 253 (AG+GG) | NA | NA | 9 | ||
| Koushik [18] | 2006 | USA | Mixed | Colorectal | PB | Taqman | 82 | 159 | 116 | 163 | 399 | 245 | 0.45 | 0.981 | 14 |
| Shrubsole [19] | 2006 | China | Asian | Breast | PB | Taqman | 621 | 393 | 70 | 687 | 422 | 76 | 0.24 | 0.304 | 14 |
| Hazra [20] | 2007 | USA | Mixed | Colorectal | PB | Taqman | 113 | 258 | 162 | 111 | 264 | 158 | 0.46 | 0.970 | 14 |
| Kim [21] | 2007 | Korea | Asian | Multiple myeloma | PB | Pyrosequencing | 91 | 69 | 14 | 857 | 718 | 125 | 0.28 | 0.127 | 11 |
| Lissowska [22] | 2007 | Poland | Caucasian | Breast | PB | PCR-RFLP | 358 | 970 | 663 | 430 | 1110 | 753 | 0.43 | 0.558 | 13 |
| Moore [23] | 2007 | Spain | Caucasian | Bladder | HB | Illumina | 267 | 531 | 291 | 232 | 510 | 274 | 0.48 | 0.857 | 10 |
| Petra [24] | 2007 | Slovenia | Caucasian | ALL | HB | PCR-RFLP | 15 | 36 | 17 | 47 | 136 | 75 | 0.45 | 0.283 | 7 |
| Suzuki [25] | 2007 | Japan | Asian | Head and neck | HB | PCR-RFLP | 108 | 100 | 29 | 332 | 315 | 64 | 0.31 | 0.382 | 9 |
| Suzuki [26] | 2007 | Japan | Asian | Lung | HB | Taqman | 235 | 256 | 54 | 484 | 446 | 100 | 0.31 | 0.852 | 9 |
| Zhang [27] | 2007 | Poland | Caucasian | Gastric | PB | Taqman | 56 | 133 | 106 | 78 | 188 | 147 | 0.42 | 0.197 | 13 |
| Bethke [28] | 2008 | Multi-center | Caucasian | Brain | PB | Illumina | 534 | 795 | 307 | 579 | 783 | 286 | 0.41 | 0.447 | 14 |
| Gra [29] b | 2008 | Russia | Caucasian | ALL | PB | PCR-based biochip | 109 (AA+AG) | 31 | 151 (AA+AG) | 95 | NA | NA | 7 | ||
| Gra [29] b | 2008 | Russia | Caucasian | AML | PB | PCR-based biochip | 26 (AA+AG) | 11 | 151 (AA+AG) | 95 | NA | NA | 7 | ||
| Gra [30] | 2008 | Russia | Caucasian | NHL | PB | PCR-based biochip | 16 | 40 | 20 | 33 | 92 | 52 | 0.45 | 0.492 | 9 |
| Gra [30] | 2008 | Russia | Caucasian | CLL | PB | PCR-based biochip | 20 | 32 | 31 | 33 | 92 | 52 | 0.45 | 0.492 | 9 |
| Ikeda [31] | 2008 | Japan | Asian | Colorectal | HB | MassARRAY | 51 | 47 | 8 | 132 | 78 | 12 | 0.23 | 0.914 | 8 |
| Ikeda [31] | 2008 | Japan | Asian | Gastric | HB | MassARRAY | 83 | 55 | 5 | 134 | 120 | 24 | 0.30 | 0.694 | 8 |
| Kim [32] | 2008 | Korea | Asian | NHL | PB | Pyrosequencing | 292 | 235 | 57 | 857 | 718 | 125 | 0.28 | 0.127 | 10 |
| Kwak [33] | 2008 | Korea | Asian | Liver | PB | PCR-RFLP | 40 | 45 | 9 | 111 | 78 | 12 | 0.25 | 0.726 | 7 |
| Lima [34] | 2008 | Brazil | Mixed | Multiple myeloma | HB | PCR-RFLP | 32 | 63 | 28 | 53 | 102 | 33 | 0.45 | 0.181 | 6 |
| Marchal [35] | 2008 | Spain | Caucasian | Prostate | HB | Real-time PCR | 38 | 105 | 39 | 46 | 111 | 47 | 0.50 | 0.207 | 8 |
| Mir [36] c | 2008 | India | Asian | Breast | HB | PCR-RFLP | 1 | 27 | 7 | 0 | 9 | 24 | 0.14 | 0.364 | 4 |
| Steck [37] | 2008 | USA | African | Colorectal | PB | Taqman | 116 | 99 | 24 | 169 | 127 | 26 | 0.28 | 0.755 | 13 |
| Steck [37] | 2008 | USA | Caucasian | Colorectal | PB | Taqman | 53 | 155 | 99 | 109 | 256 | 168 | 0.44 | 0.526 | 13 |
| Suzuki [38] | 2008 | Japan | Asian | Breast | HB | Taqman | 205 | 205 | 42 | 456 | 366 | 90 | 0.30 | 0.191 | 10 |
| Suzuki [39] | 2008 | Japan | Asian | Pancreatic | HB | Taqman | 78 | 67 | 12 | 374 | 330 | 81 | 0.31 | 0.517 | 10 |
| Theodoratou [40] | 2008 | Scotland | Caucasian | Colorectal | PB | Illumina | 200 | 456 | 339 | 198 | 482 | 329 | 0.44 | 0.370 | 12 |
| de Jonge [41] | 2009 | Netherlands | Caucasian | ALL | PB | Real-time PCR | 59 | 117 | 66 | 101 | 245 | 153 | 0.45 | 0.871 | 7 |
| Kim [42] | 2009 | Korea | Asian | ALL | PB | Pyrosequencing | 58 | 34 | 15 | 857 | 718 | 125 | 0.28 | 0.127 | 9 |
| Kim [42] | 2009 | Korea | Asian | AML | PB | Pyrosequencing | 195 | 162 | 42 | 857 | 718 | 125 | 0.28 | 0.127 | 10 |
| Kim [42] | 2009 | Korea | Asian | CML | PB | Pyrosequencing | 73 | 68 | 11 | 857 | 718 | 125 | 0.28 | 0.127 | 9 |
| Rouissi [43] | 2009 | Tunisia | African | Bladder | PB | PCR-RFLP | 59 | 88 | 38 | 77 | 85 | 29 | 0.37 | 0.490 | 5 |
| Burcos [44] c | 2010 | Romania | Caucasian | Breast | HB | PCR-RFLP | 0 | 37 | 23 | 3 | 32 | 25 | 0.32 | 0.072 | 6 |
| Burcos [44] | 2010 | Romania | Caucasian | Colorectal | HB | PCR-RFLP | 11 | 64 | 45 | 7 | 35 | 18 | 0.41 | 0.108 | 6 |
| Cai [45] | 2010 | China | Asian | Prostate | HB | PCR-RFLP | 111 | 92 | 14 | 118 | 89 | 13 | 0.26 | 0.479 | 8 |
| Eussen [46] | 2010 | Multi-center | Caucasian | Gastric | PB | MALDI-TOF MS | 58 | 100 | 81 | 156 | 286 | 165 | 0.49 | 0.157 | 12 |
| Sangrajrang [47] | 2010 | Thailand | Asian | Breast | HB | Taqman | 295 | 218 | 46 | 229 | 210 | 46 | 0.31 | 0.830 | 11 |
| Tong [48] b | 2010 | Korea | Asian | Cervical | HB | Multiplexed PCR | 137 (AA+AG) | 11 | 407 (AA+AG) | 23 | NA | NA | 9 | ||
| Wettergren [49] | 2010 | Sweden | Caucasian | Colorectal | PB | Real-time PCR | 22 | 94 | 61 | 50 | 152 | 97 | 0.42 | 0.463 | 7 |
| Curtin [50] | 2011 | USA | Mixed | Colorectal | PB | Illumina | 193 | 363 | 187 | 211 | 464 | 278 | 0.46 | 0.509 | 12 |
| Guimaraes [51] | 2011 | Brazil | Mixed | Colorectal | HB | PCR-RFLP | 26 | 55 | 32 | 53 | 102 | 33 | 0.45 | 0.181 | 6 |
| Jokic [52] | 2011 | Croatia | Caucasian | Colorectal | PB | Taqman | 53 | 159 | 88 | 74 | 143 | 83 | 0.49 | 0.428 | 10 |
| Metayer [53] | 2011 | USA | Mixed | ALL | PB | Illumina | 133 | 178 | 66 | 145 | 220 | 82 | 0.43 | 0.928 | 11 |
| Mostowska [54] | 2011 | Poland | Caucasian | Cervical | PB | HRM | 44 | 54 | 26 | 61 | 78 | 29 | 0.40 | 0.636 | 12 |
| Pardini [55] | 2011 | Czech | Caucasian | Colorectal | HB | Taqman | 113 | 330 | 218 | 291 | 671 | 410 | 0.46 | 0.592 | 11 |
| te Winkel [56] | 2011 | Netherlands | Caucasian | ALL | PB | Real-time PCR | 17 | 42 | 21 | 15 | 26 | 17 | 0.48 | 0.436 | 9 |
| Webb [57] | 2011 | Australia | Mixed | Ovarian | PB | MassARRAY | 584 | 888 | 405 | 447 | 730 | 292 | 0.44 | 0.846 | 12 |
| Weiner [58] | 2011 | Russia | Caucasian | NHL | PB | Real-time PCR | 26 | 64 | 35 | 97 | 259 | 162 | 0.44 | 0.716 | 8 |
| Yang [59] | 2011 | China | Asian | ALL | PB | Real-time PCR | 180 | 154 | 27 | 198 | 146 | 23 | 0.26 | 0.568 | 12 |
| Amigou [60] | 2012 | France | Caucasian | ALL | PB | Illumina | 112 | 187 | 110 | 95 | 226 | 120 | 0.47 | 0.553 | 13 |
| Galbiatti [61] a | 2012 | Brazil | Mixed | Head and neck | PB | Real-time PCR | 69 | 196 (AG+GG) | 149 | 317 (AG+GG) | NA | NA | 10 | ||
| Lajin [62] | 2012 | Syria | Caucasian | Breast | PB | ARMS-PCR | 40 | 59 | 20 | 43 | 58 | 25 | 0.43 | 0.499 | 4 |
| Pawlik [63] | 2012 | Poland | Caucasian | Ovarian | PB | HRM | 47 | 68 | 19 | 63 | 68 | 29 | 0.39 | 0.165 | 12 |
| Weiner [64] | 2012 | Russia | Caucasian | Breast | PB | Real-time PCR | 162 | 387 | 285 | 158 | 394 | 216 | 0.46 | 0.376 | 12 |
| Yoo [65] | 2012 | Korea | Asian | Gastric | HB | MassARRAY | 655 | 513 | 81 | 212 | 135 | 22 | 0.24 | 0.934 | 7 |
| Yoshimitsu [66] | 2012 | Japan | Asian | Colorectal | HB | PCR-RFLP | 281 | 198 | 39 | 490 | 454 | 107 | 0.32 | 0.903 | 10 |
| Yuan [67] | 2012 | China | Asian | Gastric | HB | MassARRAY | 27 | 112 | 140 | 17 | 114 | 165 | 0.25 | 0.642 | 7 |
| Chen [68] | 2013 | China | Asian | Cervical | HB | PCR-RFLP | 50 | 46 | 11 | 54 | 44 | 9 | 0.29 | 0.993 | 7 |
| Jackson [69] a | 2013 | Jamaica | African | Prostate | HB | Taqman | 111 | 84 (AG+GG) | 120 | 83 (AG+GG) | NA | NA | 7 | ||
| Liu [70] | 2013 | USA | Mixed | Colorectal | PB | Illumina | 264 | 717 | 439 | 356 | 869 | 550 | 0.45 | 0.704 | 12 |
| Morita [71] | 2013 | Japan | Asian | Colorectal | PB | PCR-RFLP | 342 | 278 | 65 | 361 | 343 | 74 | 0.32 | 0.565 | 11 |
| Tomita [72] | 2013 | Brazil | Mixed | Cervical | HB | Allele-specific PCR | 70 | 90 | 40 | 38 | 43 | 19 | 0.41 | 0.281 | 8 |
| Zhang [73] | 2013 | China | Asian | Brain | PB | PCR-RFLP | 209 | 269 | 122 | 225 | 282 | 93 | 0.39 | 0.765 | 12 |
| Chang [74] | 2014 | China | Asian | Gastric | PB | Taqman | 119 | 63 | 9 | 204 | 149 | 25 | 0.26 | 0.752 | 12 |
| Chang [74] | 2014 | China | Asian | Liver | PB | Taqman | 114 | 64 | 13 | 204 | 149 | 25 | 0.26 | 0.752 | 11 |
| Chang [74] | 2014 | China | Asian | Esophagus | PB | Taqman | 117 | 74 | 10 | 204 | 149 | 25 | 0.26 | 0.752 | 12 |
| Xu [75] | 2014 | China | Asian | Liver | HB | SNaPshot | 103 | 86 | 16 | 112 | 73 | 15 | 0.26 | 0.520 | 6 |
| Gong [76] | 2015 | USA | Caucasian | Breast | PB | Illumina | 158 | 318 | 140 | 165 | 321 | 138 | 0.48 | 0.442 | 14 |
| Greenop [77] | 2015 | Australia | Mixed | Brain | PB | MassARRAY | 80 | 148 | 90 | 102 | 264 | 175 | 0.43 | 0.890 | 11 |
| Suthandiram [78] | 2015 | Multi-center | Asian | NHL | HB | MassARRAY | 178 | 153 | 41 | 353 | 306 | 63 | 0.30 | 0.774 | 10 |
| Kim [79] | 2016 | Korea | Asian | Gastric | HB | Affymetrix Array | 136 | 111 | 23 | 295 | 211 | 35 | 0.26 | 0.739 | 10 |
| Nakao [80] | 2016 | Japan | Asian | Pancreatic | HB | Dynamic Array | 167 | 157 | 36 | 206 | 158 | 36 | 0.29 | 0.473 | 11 |
| Peres [81] | 2016 | Brazil | Mixed | Liver | HB | Real-time PCR | 12 | 50 | 9 | 105 | 179 | 72 | 0.45 | 0.787 | 8 |
| Tao [82] | 2016 | China | Asian | Breast | HB | MassARRAY | 175 | 85 | 38 | 162 | 115 | 21 | 0.26 | 0.924 | 9 |
MAF, minor allele frequency; HB: hospital based; PB: population based; NA, not applicable; PCR-RFLP: polymorphism chain reaction restriction fragment length polymorphism; MALDI-TOF MS: matrix-assisted laser desorption/ionization time-of-flight mass spectrometry; HRM: high resolution melt; ARMS-PCR: amplification refractory mutation system-PCR; ALL: acute lymphoblastic leukemia; NHL: non-Hodgkin's lymphoma; AML: acute myelogenous leukemia; CML: chronic myelogenous leukemia; CLL: chronic lymphocytic leukemia.
a Chen [17], Galbiatti [61] and Jackson [69] were only calculated for the dominant model.
b Gra [29] and Tong [48] were only calculated for the recessive model.
c Mir [36] and Burcos [44] (breast cancer) were only calculated for the recessive model and allele comparison, and the number of AA genotype was zero.
Of the 72 publications, two publications [11, 37] with different ethnic groups were separated as five independent studies and eight publications [12, 13, 29-31, 42, 44, 74] with different cancer types were also treated as 18 independent studies. For those studies [12, 13, 21, 25, 26, 29, 30, 32, 38, 39, 42, 50, 54, 63, 70, 74] with the same control group, the control numbers were calculated once in the total number. Overall, 72 publications including 85 studies of 32,272 cases and 37,427 controls were included in the final meta-analysis. Of the 85 studies, 20 studies focused on colorectal cancer [11, 14, 17, 18, 20, 31, 37, 40, 44, 49-52, 55, 66, 70, 71], ten on breast cancer [19, 22, 36, 38, 44, 47, 62, 64, 76, 82], nine on acute lymphoblastic leukemia (ALL) [13, 24, 29, 41, 42, 53, 56, 59, 60], eight on gastric cancer [12, 27, 31, 46, 65, 67, 74, 79], five on non-Hodgkin lymphoma (NHL) [13, 30, 32, 58, 78], four each on cervical cancer [48, 54, 68, 72] and liver cancer [33, 74, 75, 81], three each on prostate cancer [35, 45, 69], head and neck cancer [16, 25, 61] and brain cancer [28, 73, 77], and “other cancers” with no more than two studies. There were 37 studies on Asians, 32 studies on Caucasians, 13 studies on mixed ethnicities and three on Africans. Of all the studies, 52 were population-based and 33 were hospital-based. Furthermore, 37 studies were considered as low quality (quality score ≤ 9), and 48 studies (56.5%) were considered as high quality (quality score > 9). Controls were matched for age, sex and ethnicity in most studies.
Meta-analysis results
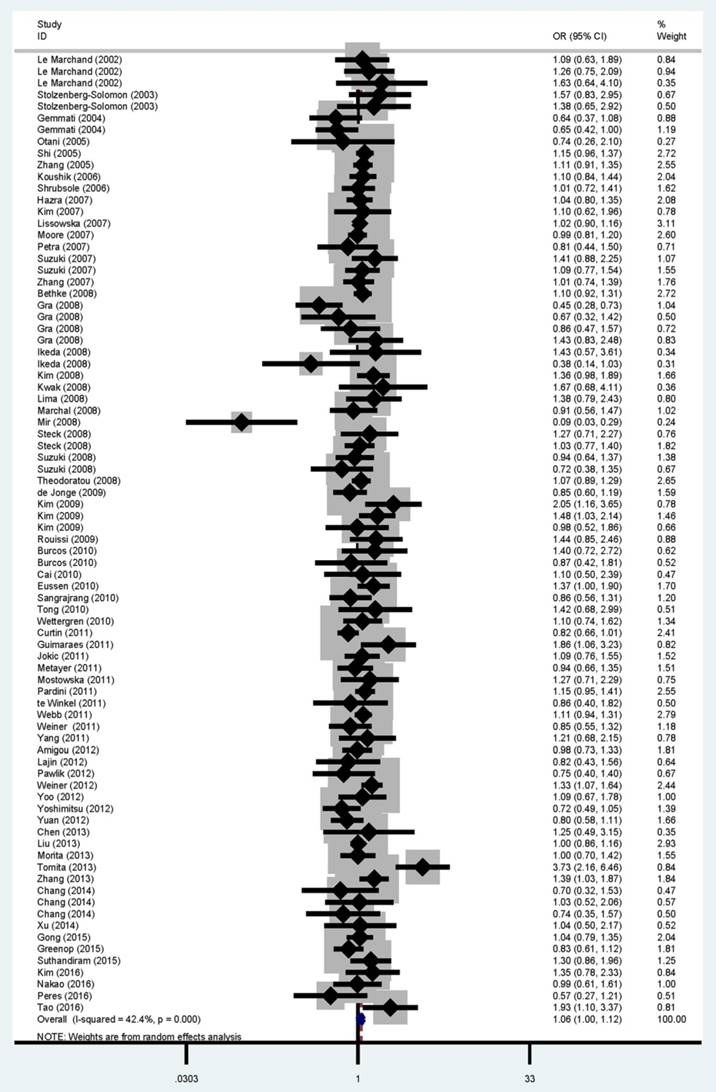
The main results of the meta-analysis are shown in Table 2 and Figure 2. Pooled analysis indicated a significant association between the MTRR A66G polymorphism and cancer risk (homozygous: OR = 1.08, 95% CI = 1.02-1.15, P = 0.009; recessive: OR = 1.06, 95% CI = 1.00-1.12, P < 0.001 and allele comparison: OR = 1.03, 95% CI = 1.00-1.06, P < 0.001). In the subgroup analysis, statistically significant associations were found for head and neck cancer (homozygous: OR = 1.49, 95% CI = 1.17-1.89, P = 0.768; dominant: OR = 1.30, 95% CI = 1.03-1.64, P = 0.143 and allele comparison: OR = 1.17, 95% CI = 1.04-1.31, P = 0.560), Caucasians (homozygous: OR = 1.09, 95% CI = 1.00-1.19, P = 0.077; dominant: OR = 1.08, 95% CI = 1.00-1.17, P = 0.045 and allele comparison: OR = 1.05, 95% CI = 1.01-1.09, P = 0.193), Africans (homozygous: OR = 1.52, 95% CI = 1.00-2.32, P = 0.577 and allele comparison: OR = 1.23, 95% CI = 1.01-1.49, P = 0.474) and high quality studies (homozygous: OR = 1.07, 95% CI = 1.00-1.15, P = 0.005 and recessive: OR = 1.06, 95% CI = 1.01-1.11, P = 0.262).
Heterogeneity and sensitivity analysis
Substantial heterogeneity was detected among all studies of the MTRR A66G polymorphism and overall cancer risk (homozygous: P = 0.009; heterozygous: P = 0.007; dominant: P = 0.001; recessive: P < 0.001 and allele comparison: P < 0.001). Therefore, the random-effects model was applied to generate wider CIs. Leave-one-out sensitivity analysis was performed and the results suggested the pooled ORs were not influenced by omitting any single study (data not shown).
Publication bias
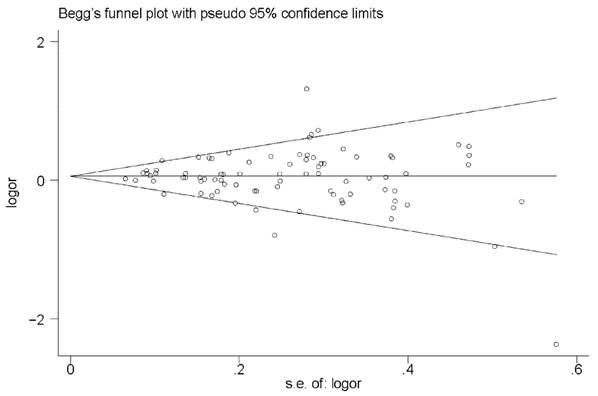
As shown by the relative symmetric funnel plot (Figure 3) and Egger's test, no evidence of publication bias was found in the current analysis under any of the models (homozygous: P = 0.913; heterozygous: P = 0.551; dominant: P = 0.510; recessive: P = 0.666 and allele comparison: P = 0.560).
FPRP test results
The significant associations were investigated using the FPRP test and the results were shown in Table 3. For a prior probability of 0.1, the FPRP value was 0.128 for the MTRR A66G polymorphism with an increased cancer risk under the homozygous model, and positive associations were also found in head and neck cancer (homozygous: FPRP = 0.017 and allele comparison: FPRP = 0.055), Caucasians (allele comparison: FPRP = 0.087) and high score studies (recessive: FPRP = 0.106). However, no positive association was found between the MTRR A66G polymorphism and cancer risk in Africans.
Meta-analysis of the association between MTRR A66G polymorphism and cancer risk.
| Variables | No. of studies | Sample size (case/controls) | Homozygous | Heterozygous | Recessive | Dominant | Allele comparison | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG vs. AA | AG vs. AA | GG vs. (AA + AG) | (GG + AG) vs. AA | G vs. A | ||||||||
| OR (95% CI) | Phet | OR (95% CI) | Phet | OR (95% CI) | Phet | OR (95% CI) | Phet | OR (95% CI) | Phet | |||
| All a | 85 | 32,272/37,427 | 1.08 (1.02-1.15) | 0.009 | 1.01 (0.97-1.06) | 0.007 | 1.06 (1.00-1.12) | <0.001 | 1.04 (0.99-1.08) | 0.001 | 1.03 (1.00-1.06) | <0.001 |
| Cancer type | ||||||||||||
| Colorectal | 20 | 8,057/10,465 | 1.09 (0.96-1.25) | 0.031 | 1.05 (0.95-1.16) | 0.030 | 1.04 (0.97-1.11) | 0.462 | 1.07 (0.97-1.19) | 0.006 | 1.05 (0.98-1.12) | 0.007 |
| Breast | 10 | 6,048/5,872 | 1.08 (0.96-1.21) | 0.488 | 0.99 (0.89-1.11) | 0.131 | 0.99 (0.81-1.22) | 0.001 | 1.02 (0.94-1.11) | 0.362 | 1.01 (0.92-1.11) | 0.018 |
| ALL | 9 | 1,893/3,770 | 0.90 (0.72-1.13) | 0.228 | 0.88 (0.76-1.03) | 0.367 | 0.89 (0.70-1.14) | 0.013 | 0.89 (0.78-1.02) | 0.472 | 0.93 (0.85-1.02) | 0.547 |
| Gastric | 8 | 2,756/2,504 | 0.96 (0.72-1.29) | 0.054 | 0.95 (0.80-1.12) | 0.159 | 1.02 (0.82-1.27) | 0.109 | 0.94 (0.78-1.14) | 0.041 | 0.97 (0.84-1.12) | 0.010 |
| NHL | 5 | 1,357/1,674 | 1.00 (0.74-1.35) | 0.126 | 0.97 (0.84-1.11) | 0.998 | 0.99 (0.74-1.33) | 0.053 | 0.99 (0.87-1.13) | 0.911 | 0.99 (0.89-1.11) | 0.295 |
| Cervical | 4 | 579/805 | 1.22 (0.80-1.86) | 0.968 | 1.07 (0.78-1.46) | 0.882 | 1.77 (0.98-3.20) | 0.029 | 1.11 (0.83-1.48) | 0.945 | 1.10 (0.90-1.36) | 0.982 |
| Liver | 4 | 561/757 | 1.19 (0.79-1.78) | 0.600 | 1.33 (0.84-2.10) | 0.011 | 0.97 (0.65-1.45) | 0.335 | 1.29 (0.86-1.94) | 0.022 | 1.11 (0.89-1.38) | 0.151 |
| Brain | 3 | 2,554/2,789 | 1.05 (0.72-1.52) | 0.009 | 0.98 (0.79-1.21) | 0.091 | 1.08 (0.84-1.40) | 0.054 | 0.99 (0.77-1.27) | 0.029 | 1.02 (0.85-1.22) | 0.014 |
| Head and neck | 3 | 1,223/1,700 | 1.49 (1.17-1.89) | 0.768 | 1.24 (0.79-1.94) | 0.025 | 1.15 (0.96-1.38) | 0.346 | 1.30 (1.03-1.64) | 0.143 | 1.17 (1.04-1.31) | 0.560 |
| Prostate | 3 | 594/627 | 1.05 (0.65-1.71) | 0.798 | 1.12 (0.82-1.52) | 0.899 | 0.96 (0.64-1.44) | 0.689 | 1.10 (0.87-1.40) | 0.999 | 1.04 (0.84-1.27) | 0.718 |
| Other cancers | 16 | 6,650/6,464 | 1.14 (1.01-1.28) | 0.282 | 1.01 (0.94-1.10) | 0.335 | 1.10 (1.01-1.20) | 0.533 | 1.06 (0.97-1.15) | 0.211 | 1.06 (1.00-1.11) | 0.340 |
| Ethnicity | ||||||||||||
| Asian | 37 | 11,829/13,248 | 1.11 (0.99-1.24) | 0.080 | 0.98 (0.92-1.05) | 0.063 | 1.09 (0.97-1.22) | 0.006 | 1.01 (0.95-1.08) | 0.019 | 1.02 (0.97-1.08) | 0.001 |
| Caucasian | 32 | 13,351/16,506 | 1.09 (1.00-1.19) | 0.077 | 1.08 (0.99-1.16) | 0.078 | 1.03 (0.96-1.09) | 0.144 | 1.08 (1.00-1.17) | 0.045 | 1.05 (1.01-1.09) | 0.193 |
| African | 3 | 619/716 | 1.52 (1.00-2.32) | 0.577 | 1.21 (0.92-1.60) | 0.553 | 1.36 (0.92-2.02) | 0.751 | 1.21 (0.97-1.51) | 0.624 | 1.23 (1.01-1.49) | 0.474 |
| Mixed | 13 | 6,473/6,957 | 1.01 (0.88-1.15) | 0.084 | 0.96 (0.86-1.06) | 0.184 | 1.12 (0.96-1.32) | <0.001 | 1.00 (0.90-1.11) | 0.075 | 1.01 (0.94-1.07) | 0.088 |
| Source of control | ||||||||||||
| PB | 52 | 21,300/24,134 | 1.06 (0.99-1.14) | 0.087 | 0.99 (0.94-1.04) | 0.304 | 1.05 (0.99-1.11) | 0.037 | 1.01 (0.97-1.06) | 0.135 | 1.02 (0.99-1.06) | 0.075 |
| HB | 33 | 10,972/13,293 | 1.12 (0.99-1.26) | 0.019 | 1.06 (0.97-1.16) | 0.002 | 1.07 (0.94-1.21) | <0.001 | 1.08 (0.99-1.18) | 0.001 | 1.04 (0.98-1.11) | <0.001 |
| Score | ||||||||||||
| Low | 37 | 6,610/9,768 | 1.13 (0.99-1.29) | 0.265 | 1.05 (0.96-1.16) | 0.144 | 1.06 (0.90-1.24) | 0.000 | 1.08 (0.99-1.17) | 0.299 | 1.05 (0.98-1.12) | 0.042 |
| High | 48 | 25,662/27,659 | 1.07 (1.00-1.15) | 0.005 | 1.00 (0.95-1.05) | 0.010 | 1.06 (1.01-1.11) | 0.262 | 1.02 (0.97-1.08) | <0.001 | 1.02 (0.99-1.06) | 0.001 |
Het, heterogeneity; ALL: acute lymphoblastic leukemia; NHL: non-Hodgkin's lymphoma; PB: population based; HB: hospital based.
a The number of controls was only calculated once if the same controls were used.
Forest plot for overall cancer risk associated with the MTRR A66G polymorphism by a recessive model. For each study, the estimated OR and its 95% CI are plotted with a box and a horizontal line. ¯, pooled ORs and its 95% CIs.
Funnel plot for the MTRR A66G polymorphism and cancer risk by a recessive model.
False-positive report probability values for associations between cancer risk and genotypes of MTRR A66G polymorphism.
| Genotype | Crude OR (95% CI) | P-value a | Statistical Power b | Prior probability | ||||
|---|---|---|---|---|---|---|---|---|
| 0.25 | 0.1 | 0.01 | 0.001 | 0.0001 | ||||
| All patients | ||||||||
| Homozygous | 1.08 (1.02-1.15) | 0.016 | 1.000 | 0.047 | 0.128 | 0.618 | 0.942 | 0.994 |
| Recessive | 1.06 (1.00-1.12) | 0.038 | 1.000 | 0.102 | 0.255 | 0.790 | 0.974 | 0.997 |
| Allele comparison | 1.03 (1.00-1.06) | 0.044 | 1.000 | 0.116 | 0.282 | 0.812 | 0.978 | 0.998 |
| Cancer type-head and neck cancer | ||||||||
| Homozygous | 1.49 (1.17-1.89) | 0.001 | 0.522 | 0.006 | 0.017 | 0.161 | 0.660 | 0.951 |
| Dominant | 1.30 (1.03-1.64) | 0.027 | 0.886 | 0.083 | 0.214 | 0.750 | 0.968 | 0.997 |
| Allele comparison | 1.17 (1.04-1.31) | 0.006 | 1.000 | 0.019 | 0.055 | 0.391 | 0.886 | 0.985 |
| Ethnicity-Caucasian | ||||||||
| Homozygous | 1.09 (1.00-1.19) | 0.054 | 1.000 | 0.140 | 0.328 | 0.843 | 0.982 | 0.998 |
| Dominant | 1.08 (1.00-1.17) | 0.059 | 1.000 | 0.151 | 0.349 | 0.885 | 0.983 | 0.998 |
| Allele comparison | 1.05 (1.01-1.09) | 0.010 | 1.000 | 0.031 | 0.087 | 0.511 | 0.913 | 0.991 |
| Ethnicity-African | ||||||||
| Homozygous | 1.52 (1.00-2.32) | 0.052 | 0.476 | 0.248 | 0.497 | 0.916 | 0.991 | 0.999 |
| Allele comparison | 1.23 (1.01-1.49) | 0.034 | 0.979 | 0.095 | 0.240 | 0.777 | 0.972 | 0.997 |
| Score-high | ||||||||
| Homozygous | 1.07 (1.00-1.15) | 0.066 | 1.000 | 0.165 | 0.372 | 0.867 | 0.985 | 0.998 |
| Recessive | 1.06 (1.01-1.11) | 0.013 | 1.000 | 0.038 | 0.106 | 0.567 | 0.930 | 0.992 |
aChi-square test was used to calculate the genotype frequency distributions.
bStatistical power was calculated using the number of observations in the subgroup and the OR and P values in this table.
Discussion
Folate is a critical coenzyme in DNA synthesis, and the maintenance of methylation, and folate deficiency has been reported to be associated with various human malignancies [113, 114]. MTRR plays a key role in folate-dependent homocysteine remethylation and is required in the regulation of MTR activity. The A66G polymorphism is one of the most common polymorphisms in the MTRR gene, which was first reported in 1998 [115], and the variant enzyme has reduced affinity for MTR [116]. The reported associations between the MTRR A66G polymorphism and cancer susceptibility are inconsistent due to the small sample sizes in individual studies, ethnic differences and research methodology.
Our present study represents an updated comprehensive meta-analysis of the association between the MTRR A66G polymorphism and cancer risk and included 85 studies with 32,272 cases and 37,427 controls. The results revealed that the MTRR A66G polymorphism was significantly associated with an increased overall cancer risk. In the subgroup analysis, the association was more evident for head and neck cancer, Caucasians, Africans and high quality studies. However, the results for Africans need further validation due to the high probability of false-positive reports. Furthermore, no potential publication bias was detected by the funnel plot and Egger's regression test, indicating the robustness of the results in this study.
One previous meta-analysis focused on the MTRR A66G polymorphism and overall cancer risk. In the meta-analysis by Han et al. [117], which included 35 studies with 18,661 cases and 27,678 controls, an increased overall cancer risk was observed only under the allele comparison and homozygous model. In the subgroup analysis, significantly increased risks were found in Asians. We found this polymorphism to be associated with an increased overall risk also under the recessive model and increased cancer risks in head and neck cancer, Caucasians and Africans, but not in Asians, which were different from the previous meta-analysis; this result presumably occurred because our analysis was based on a much larger sample size, thereby increasing the statistical power. In the subgroup analysis by cancer type, we did not find any significant association between the MTRR A66G polymorphism and colorectal cancer in any comparison models, a finding that was inconsistent with previous meta-analyses [6, 118]. The discrepancy occurred because, in the current study, we added many recently published studies and even included several Chinese publications, allowing the more precise detection of an association.
Large and well-designed studies with “statistically significant” results for genetic variants turned out to be false-positive findings [119, 120]. Thus, we used the FPRP test to investigate positive associations in the current meta-analysis. Interestingly, the FPRP test results showed that the MTRR A66G polymorphism could actually increase cancer susceptibility. In the subgroup analysis, the FPRP test indicated that the MTRR A66G polymorphism increased cancer susceptibility in head and neck cancer, Caucasians and high score studies. The significant association with Africans in the present meta-analysis was false positive, which may due to the limited sample size.
Although we conducted a comprehensive literature search and included the latest studies on the MTRR A66G polymorphism and cancer risk, some possible limitations in this meta-analysis should be addressed. First, the number of cases in the individual studies was small (<1000) in all but eight studies [15, 19, 22, 23, 28, 57, 65, 70]; this limitation may affect the investigation of the real association. Second, our results were based on unadjusted estimates, so the estimates were relatively imprecise. Third, the effects of gene-gene, and gene-environment interactions were not evaluated due to the lack of original data, which may affect cancer risk. Fourth, in the subgroup analysis, only three studies were carried out in Africans, which may lead to relatively weak power to detect the real association. Finally, only studies published in English and Chinese were included, so we may have missed publications in other languages.
In conclusion, we performed this updated meta-analysis with the latest published studies and obtained a more precise estimation of the association between the MTRR A66G polymorphism and cancer risk. However, it is necessary to conduct well-designed prospective studies with larger sample sizes to verify our findings.
Acknowledgements
This work was supported by the Key Scientific and Technological Project of Henan province (Grant No. 162102310413) and the Special Financial Grant from the China Postdoctoral Science Fundation (Grant No. 2014T70836).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108
2. Wiseman M. The second World Cancer Research Fund/American Institute for Cancer Research expert report. Food, nutrition, physical activity, and the prevention of cancer: a global perspective. Proc Nutr Soc. 2008;67:253-256
3. Sanjoaquin MA, Allen N, Couto E, Roddam AW, Key TJ. Folate intake and colorectal cancer risk: a meta-analytical approach. Int J Cancer. 2005;113:825-828
4. Lajous M, Lazcano-Ponce E, Hernandez-Avila M, Willett W, Romieu I. Folate, vitamin B6, and vitamin B12 intake and the risk of breast cancer among Mexican women. Cancer Epidemiol Biomarkers Prev. 2006;15:443-448
5. Shen H, Wei Q, Pillow PC, Amos CI, Hong WK, Spitz MR. Dietary folate intake and lung cancer risk in former smokers a case-control analysis. Cancer Epidemiol Biomarkers Prev. 2003;12:980-986
6. Zhou D, Mei Q, Luo H, Tang B, Yu P. The polymorphisms in methylenetetrahydrofolate reductase, methionine synthase, methionine synthase reductase, and the risk of colorectal cancer. Int J Biol Sci. 2012;8:819-830
7. Kim YI. Methylenetetrahydrofolate reductase polymorphisms, folate, and cancer risk: a paradigm of gene-nutrient interactions in carcinogenesis. Nutr Rev. 2000;58:205-209
8. Stempak JM, Sohn KJ, Chiang EP, Shane B, Kim YI. Cell and stage of transformation-specific effects of folate deficiency on methionine cycle intermediates and DNA methylation in an in vitro model. Carcinogenesis. 2005;26:981-990
9. Fang DH, Ji Q, Fan CH, An Q, Li J. Methionine synthase reductase A66G polymorphism and leukemia risk: evidence from published studies. Leuk Lymphoma. 2014;55:1910-1914
10. Gaughan DJ, Kluijtmans LA, Barbaux S, McMaster D, Young IS, Yarnell JW. et al. The methionine synthase reductase (MTRR) A66G polymorphism is a novel genetic determinant of plasma homocysteine concentrations. Atherosclerosis. 2001;157:451-456
11. Le Marchand L, Donlon T, Hankin JH, Kolonel LN, Wilkens LR, Seifried A. B-vitamin intake, metabolic genes, and colorectal cancer risk (United States). Cancer Causes Control. 2002;13:239-248
12. Stolzenberg-Solomon RZ, Qiao YL, Abnet CC, Ratnasinghe DL, Dawsey SM, Dong ZW. et al. Esophageal and gastric cardia cancer risk and folate- and vitamin B12-related polymorphisms in Linxian, China. Cancer Epidemiol Biomarkers Prev. 2003;12:1222-1226
13. Gemmati D, Ongaro A, Scapoli GL, Della Porta M, Tognazzo S, Serino ML. et al. Common gene polymorphisms in the metabolic folate and methylation pathway and the risk of acute lymphoblastic leukemia and non-Hodgkin's lymphoma in adults. Cancer Epidemiol Biomarkers Prev. 2004;13:787-794
14. Otani T, Iwasaki M, Hanaoka T, Kobayashi M, Ishihara J, Natsukawa S. et al. Folate, vitamin B6, vitamin B12, and vitamin B2 intake, genetic polymorphisms of related enzymes, and risk of colorectal cancer in a hospital-based case-control study in Japan. Nutr Cancer. 2005;53:42-50
15. Shi Q, Zhang Z, Li G, Pillow PC, Hernandez LM, Spitz MR. et al. Polymorphisms of methionine synthase and methionine synthase reductase and risk of lung cancer: A case-control analysis. Pharmacogenet Genomics. 2005;15:547-555
16. Zhang Z, Shi Q, Liu Z, Sturgis EM, Spitz MR, Wei Q. Polymorphisms of methionine synthase and methionine synthase reductase and risk of squamous cell carcinoma of the head and neck: A case-control analysis. Cancer Epidemiol Biomarkers Prev. 2005;14:1188-1193
17. Chen K, Song L, Jin MJ, Fan CH, Jiang QT, Yu WP. [Association between genetic polymorphisms in folate metabolic enzyme genes and colorectal cancer: a nested case-control study]. Zhonghua Zhong Liu Za Zhi. 2006;28:429-432
18. Koushik A, Kraft P, Fuchs CS, Hankinson SE, Willett WC, Giovannucci EL. et al. Nonsynonymous polymorphisms in genes in the one-carbon metabolism pathway and associations with colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:2408-2417
19. Shrubsole MJ, Gao YT, Cai Q, Shu XO, Dai Q, Jin F. et al. MTR and MTRR polymorphisms, dietary intake, and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2006;15:586-588
20. Hazra A, Wu K, Kraft P, Fuchs CS, Giovannucci EL, Hunter DJ. Twenty-four non-synonymous polymorphisms in the one-carbon metabolic pathway and risk of colorectal adenoma in the Nurses' Health Study. Carcinogenesis. 2007;28:1510-1519
21. Kim HN, Kim YK, Lee IK, Lee JJ, Yang DH, Park KS. et al. Polymorphisms involved in the folate metabolizing pathway and risk of multiple myeloma. Am J Hematol. 2007;82:798-801
22. Lissowska J, Gaudet MM, Brinton LA, Chanock SJ, Peplonska B, Welch R. et al. Genetic polymorphisms in the one-carbon metabolism pathway and breast cancer risk: A population-based case-control study and meta-analyses. Int J Cancer. 2007;120:2696-2703
23. Moore LE, Malats N, Rothman N, Real FX, Kogevinas M, Karami S. et al. Polymorphisms in one-carbon metabolism and trans-sulfuration pathway genes and susceptibility to bladder cancer. Int J Cancer. 2007;120:2452-2458
24. Petra BG, Janez J, Vita D. Gene-gene interactions in the folate metabolic pathway influence the risk for acute lymphoblastic leukemia in children. Leuk Lymphoma. 2007;48:786-792
25. Suzuki T, Matsuo K, Hasegawa Y, Hiraki A, Wakai K, Hirose K. et al. One-carbon metabolism-related gene polymorphisms and risk of head and neck squamous cell carcinoma: Case-control study. Cancer Sci. 2007;98:1439-1446
26. Suzuki T, Matsuo K, Hiraki A, Saito T, Sato S, Yatabe Y. et al. Impact of one-carbon metabolism-related gene polymorphisms on risk of lung cancer in Japan: a case control study. Carcinogenesis. 2007;28:1718-1725
27. Zhang FF, Terry MB, Hou L, Chen J, Lissowska J, Yeager M. et al. Genetic polymorphisms in folate metabolism and the risk of stomach cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:115-121
28. Bethke L, Webb E, Murray A, Schoemaker M, Feychting M, Lonn S. et al. Functional polymorphisms in folate metabolism genes influence the risk of meningioma and glioma. Cancer Epidemiol Biomarkers Prev. 2008;17:1195-1202
29. Gra OA, Glotov AS, Kozhekbaeva Z, Makarova OV, Nasedkina TV. Genetic polymorphism in GST, NAT2, and MTRR and susceptibility to childhood acute leukemia. Mol Biol. 2008;42:214-225
30. Gra OA, Glotov AS, Nikitin EA, Glotov OS, Kuznetsova VE, Chudinov AV. et al. Polymorphisms in xenobiotic-metabolizing genes and the risk of chronic lymphocytic leukemia and non-Hodgkin's lymphoma in adult Russian patients. Am J Hematol. 2008;83:279-287
31. Ikeda S, Sasazuki S, Natsukawa S, Shaura K, Koizumi Y, Kasuga Y. et al. Screening of 214 single nucleotide polymorphisms in 44 candidate cancer susceptibility genes: a case-control study on gastric and colorectal cancers in the Japanese population. Am J Gastroenterol. 2008;103:1476-1487
32. Kim HN, Lee IK, Kim YK, Tran HTT, Yang DH, Lee JJ. et al. Association between folate-metabolizing pathway polymorphism and non-Hodgkin lymphoma. Br J Haemato. 2008;140:287-294
33. Kwak SY, Kim UK, Cho HJ, Lee HK, Kim HJ, Kim NK. et al. Methylenetetrahydrofolate reductase (MTHFR) and methionine synthase reductase (MTRR) gene polymorphisms as risk factors for hepatocellular carcinoma in a Korean population. Anticancer Res. 2008;28:2807-2811
34. Lima CS, Ortega MM, Ozelo MC, Araujo RC, De Souza CA, Lorand-Metze I. et al. Polymorphisms of methylenetetrahydrofolate reductase (MTHFR), methionine synthase (MTR), methionine synthase reductase (MTRR), and thymidylate synthase (TYMS) in multiple myeloma risk. Leuk Res. 2008;32:401-405
35. Marchal C, Redondo M, Reyes-Engel A, Perea-Milla E, Gaitan M, Machuca J. et al. Association between polymorphisms of folate-metabolizing enzymes and risk of prostate cancer. Eur J Surg Oncol. 2008;34:805-810
36. Mir MM, Dar JA, Dar NA, Dar MS, Salam I, Lone MM. et al. Combined impact of polymorphism of folate metabolism genes; glutamate carboxypeptidase, methylene tetrahydrofolate reductase and methionine synthase reductase on breast cancer susceptibility in Kashmiri women. Int J Health Sci. 2008;2:3-14
37. Steck SE, Keku T, Butler LM, Galanko J, Massa B, Millikan RC. et al. Polymorphisms in methionine synthase, methionine synthase reductase and serine hydroxymethyltransferase, folate and alcohol intake, and colon cancer risk. J Nutrigenet Nutrigenomics. 2008;1:196-204
38. Suzuki T, Matsuo K, Hirose K, Hiraki A, Kawase T, Watanabe M. et al. One-carbon metabolism-related gene polymorphisms and risk of breast cancer. Carcinogenesis. 2008;29:356-362
39. Suzuki T, Matsuo K, Sawaki A, Mizuno N, Hiraki A, Kawase T. et al. Alcohol drinking and one-carbon metabolism-related gene polymorphisms on pancreatic cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17:2742-2747
40. Theodoratou E, Farrington SM, Tenesa A, McNeill G, Cetnarskyj R, Barnetson RA. et al. Dietary vitamin B6 intake and the risk of colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:171-182
41. de Jonge R, Tissing WJ, Hooijberg JH, Jansen G, Kaspers GJ, Lindemans J. et al. Polymorphisms in folate-related genes and risk of pediatric acute lymphoblastic leukemia. Blood. 2009;113:2284-2289
42. Kim HN, Kim YK, Lee IK, Yang DH, Lee JJ, Shin MH. et al. Association between polymorphisms of folate-metabolizing enzymes and hematological malignancies. Leukemia Res. 2009;33:82-87
43. Rouissi K, Ouerhani S, Oliveira E, Marrakchi R, Cherni L, Othman FB. et al. Polymorphisms in one-carbon metabolism pathway genes and risk for bladder cancer in a Tunisian population. Cancer Genet Cytogen. 2009;195:43-53
44. Burcoæ T, Toma M, Stavarachi M, Cimponeriu D, Apostol P, Popa E. et al. MTRR polymorphism and the risk for colorectal and breast cancer in Romanian patients-a preliminary study. Chirurgia. 2010;105:379-382
45. Cai D, Ning L, Pan C, Liu X, Bu R, Chen X. et al. Association of polymorphisms in folate metabolic genes and prostate cancer risk: A case-control study in a Chinese population. J Genet. 2010;89:263-267
46. Eussen SJPM, Vollset SE, Hustad S, Midttun Ø, Meyer K, Fredriksen Å. et al. Vitamins B2 and B6 and genetic polymorphisms related to one-carbon metabolism as risk factors for gastric adenocarcinoma in the European prospective investigation into cancer and nutrition. Cancer Epidemiol Biomarkers Prev. 2010;19:28-38
47. Sangrajrang S, Sato Y, Sakamoto H, Ohnami S, Khuhaprema T, Yoshida T. Genetic polymorphisms in folate and alcohol metabolism and breast cancer risk: A case-control study in Thai women. Breast Cancer Res Treat. 2010;123:885-893
48. Tong SY, Lee JM, Song ES, Lee KB, Kim MK, Yun YM. et al. The effects of polymorphisms in methylenetetrahydrofolate reductase (MTHFR), methionine synthase (MTR), and methionine synthase reductase (MTRR) on the risk of cervical intraepithelial neoplasia and cervical cancer in Korean women. Cancer Causes Control. 2010;21:23-30
49. Wettergren Y, Odin E, Carlsson G, Gustavsson B. MTHFR, MTR, and MTRR polymorphisms in relation to p16INK4A hypermethylation in mucosa of patients with colorectal cancer. Mol Med. 2010;16:425-432
50. Curtin K, Ulrich CM, Samowitz WS, Wolff RK, Duggan DJ, Makar KW. et al. Candidate pathway polymorphisms in one-carbon metabolism and risk of rectal tumor mutations. Int J Mol Epidemiol Genet. 2011;2:1-8
51. Guimaraes JL, Ayrizono Mde L, Coy CS, Lima CS. Gene polymorphisms involved in folate and methionine metabolism and increased risk of sporadic colorectal adenocarcinoma. Tumor Biol. 2011;32:853-861
52. Jokić M, Brčić-Kostić K, Stefulj J, Ivković TC, Božo L, Gamulin M. et al. Association of MTHFR, MTR, MTRR, RFC1, and DHFR gene polymorphisms with susceptibility to sporadic colon cancer. DNA Cell Biol. 2011;30:771-776
53. Metayer C, Scelo G, Chokkalingam AP, Barcellos LF, Aldrich MC, Chang JS. et al. Genetic variants in the folate pathway and risk of childhood acute lymphoblastic leukemia. Cancer Causes Control. 2011;22:1243-1258
54. Mostowska A, Myka M, Lianeri M, Roszak A, Jagodzinski PP. Folate and choline metabolism gene variants and development of uterine cervical carcinoma. Clin Biochem. 2011;44:596-600
55. Pardini B, Kumar R, Naccarati A, Prasad RB, Forsti A, Polakova V. et al. MTHFR and MTRR genotype and haplotype analysis and colorectal cancer susceptibility in a case-control study from the Czech Republic. Mutat Res-Gen Tox En. 2011;721:74-80
56. te Winkel ML, de Muinck Keizer-Schrama SM, de Jonge R, van Beek RD, van der Sluis IM, Hop WC. et al. Germline variation in the MTHFR and MTRR genes determines the nadir of bone density in pediatric acute lymphoblastic leukemia: a prospective study. Bone. 2011;48:571-577
57. Webb PM, Ibiebele TI, Hughes MC, Beesley J, Van Der Pols JC, Chen X. et al. Folate and related micronutrients, folate-metabolising genes and risk of ovarian cancer. Eur J Clin Nutr. 2011;65:1133-1140
58. Weiner AS, Beresina OV, Voronina EN, Voropaeva EN, Boyarskih UA, Pospelova TI. et al. Polymorphisms in folate-metabolizing genes and risk of non-Hodgkin's lymphoma. Leuk Res. 2011;35:508-515
59. Yang L, Liu L, Wang J, Qiu L, Mi Y, Ma X. et al. Polymorphisms in folate-related genes: Impact on risk of adult acute lymphoblastic leukemia rather than pediatric in Han Chinese. Leuk Lymphoma. 2011;52:1770-1776
60. Amigou A, Rudant J, Orsi L, Goujon-Bellec S, Leverger G, Baruchel A. et al. Folic acid supplementation, MTHFR and MTRR polymorphisms, and the risk of childhood leukemia: the ESCALE study (SFCE). Cancer Causes Control. 2012;23:1265-1277
61. Galbiatti AL, da Silva LM, Ruiz-Cintra MT, Raposo LS, Maniglia JV, Pavarino EC. et al. Association between 11 genetic polymorphisms in folate-metabolising genes and head and neck cancer risk. Eur J Cancer. 2012;48:1525-1531
62. Lajin B, Alhaj Sakur A, Ghabreau L, Alachkar A. Association of polymorphisms in one-carbon metabolizing genes with breast cancer risk in Syrian women. Tumor Biol. 2012;33:1133-1139
63. Pawlik P, Mostowska A, Lianeri M, Sajdak S, Kedzia H, Jagodzinski PP. Folate and choline metabolism gene variants in relation to ovarian cancer risk in the Polish population. Mol Biol Rep. 2012;39:5553-5560
64. Weiner AS, Boyarskikh UA, Voronina EN, Selezneva IA, Sinkina TV, Lazarev AF. et al. Polymorphisms in the folate-metabolizing genes MTR, MTRR, and CBS and breast cancer risk. Cancer Epidemiol. 2012;36:e95-e100
65. Yoo JY, Kim SY, Hwang JA, Hong SH, Shin A, Choi IJ. et al. Association study between folate pathway gene single nucleotide polymorphisms and gastric cancer in Koreans. Genomics Inform. 2012;10:184-193
66. Yoshimitsu S, Morita M, Hamachi T, Tabata S, Abe H, Tajima O. et al. Methionine synthase and thymidylate synthase gene polymorphisms and colorectal adenoma risk: the self defense forces study. Mol Carcinog. 2012;51:E151-E157
67. Yuan LJ, Jin TB, Yin JK, Du XL, Wang Q, Dong R. et al. Polymorphisms of tumor-related genes IL-10, PSCA, MTRR and NOC3L are associated with the risk of gastric cancer in the Chinese Han population. Cancer Epidemiol. 2012;36:e366-e372
68. Chen F, Wang JT, Ding L, Zhou Q, Wu YY. [Role of serum folate, polymorphisms related folate carrier gene and methionine synthase reductase gene in cervical cancer]. Zhong Liu Yan Jiu Yu Lin Chuang. 2013;25:437-440
69. Jackson MD, Tulloch-Reid MK, McFarlane-Anderson N, Watson A, Seers V, Bennett FI. et al. Complex interaction between serum folate levels and genetic polymorphisms in folate pathway genes: Biomarkers of prostate cancer aggressiveness. Genes Nutr. 2013;8:199-207
70. Liu AY, Scherer D, Poole E, Potter JD, Curtin K, Makar K. et al. Gene-diet-interactions in folate-mediated one-carbon metabolism modify colon cancer risk. Mol Nutr Food Res. 2013;57:721-734
71. Morita M, Yin G, Yoshimitsu S, Ohnaka K, Toyomura K, Kono S. et al. Folate-related nutrients, genetic polymorphisms, and colorectal cancer risk: the fukuoka colorectal cancer study. Asian Pac J Cancer Prev. 2013;14:6249-6256
72. Tomita LY, D'Almeida V, Villa LL, Franco EL, Cardoso MA, Group BS. Polymorphisms in genes involved in folate metabolism modify the association of dietary and circulating folate and vitamin B-6 with cervical neoplasia. J Nutr. 2013;143:2007-2014
73. Zhang J, Zhou YW, Shi HP, Wang YZ, Li GL, Yu HT. et al. 5,10-Methylenetetrahydrofolate reductase (MTHFR), methionine synthase (MTRR), and methionine synthase reductase (MTR) gene polymorphisms and adult meningioma risk. J Neurooncol. 2013;115:233-239
74. Chang SC, Chang PY, Butler B, Goldstein BY, Mu L, Cai L. et al. Single nucleotide polymorphisms of one-carbon metabolism and cancers of the esophagus, stomach, and liver in a Chinese population. PLoS One. 2014;9:e109235
75. Xu MJ, Gu ZY, Zhao JY, Wang HY, Liu J, Chen Y. [Association study on genetic polymorphisms of folate metabolism genes and susceptibility of hepatocelluar carcinoma]. Fudan Xue Bao Zi Ran Ke Xue Ban. 2014;53:716-723
76. Gong Z, Yao S, Zirpoli G, David Cheng TY, Roberts M, Khoury T. et al. Genetic variants in one-carbon metabolism genes and breast cancer risk in European American and African American women. Int J Cancer. 2015;137:666-677
77. Greenop KR, Scott RJ, Attia J, Bower C, de Klerk NH, Norris MD. et al. Folate pathway gene polymorphisms and risk of childhood brain tumors: results from an Australian case-control study. Cancer Epidemiol Biomarkers Prev. 2015;24:931-937
78. Suthandiram S, Gan GG, Mohd Zain S, Bee PC, Lian LH, Chang KM. et al. Genetic polymorphisms in the one-carbon metabolism pathway genes and susceptibility to non-Hodgkin lymphoma. Tumor Biol. 2015;36:1819-1834
79. Kim W, Woo HD, Lee J, Choi IJ, Kim YW, Sung J. et al. Dietary folate, one-carbon metabolism-related genes, and gastric cancer risk in Korea. Mol Nutr Food Res. 2016;60:337-345
80. Nakao H, Wakai K, Ishii N, Kobayashi Y, Ito K, Yoneda M. et al. Associations between polymorphisms in folate-metabolizing genes and pancreatic cancer risk in Japanese subjects. BMC Gastroenterol. 2016;16:83
81. Peres NP, Galbiatti-Dias ALS, Silva RFd, Pavarino ÉC, Goloni-Bertollo EM, Ruiz-Cintra MT. Polymorphisms of folate metabolism genes in patients with cirrhosis and hepatocellular carcinoma. World J Hepatol. 2016;8:1234-1243
82. Tao SL, Wang H, He BS, Xu Y, Zhu F. [Association of single nucleotide polymorphisms of folate metabolism-related enzyme gene and risk of breast cancer]. Acta Universitatis Medicinalis Nanjing (Natural science). 2016;4:473-478
83. He J, Liao XY, Zhu JH, Xue WQ, Shen GP, Huang SY. et al. Association of MTHFR C677T and A1298C polymorphisms with non-Hodgkin lymphoma susceptibility: evidence from a meta-analysis. Sci Rep. 2014;4:6159
84. Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies. J Natl Cancer Inst. 1959;22:719-748
85. DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177-188
86. Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088-1101
87. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. Br Med J. 1997;315:629-634
88. Wacholder S, Chanock S, Garciaclosas M, Ghormli LE, Rothman N. Assessing the probability that a positive report is false: An approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96:434-442
89. Matakidou A, El Galta R, Rudd MF, Webb EL, Bridle H, Eisen T. et al. Prognostic significance of folate metabolism polymorphisms for lung cancer. Br J Cancer. 2007;97:247-252
90. Jin G, Huang J, Hu Z, Dai J, Tang R, Chen Y. et al. Genetic variants in one-carbon metabolism-related genes contribute to NSCLC prognosis in a Chinese population. Cancer. 2010;116:5700-5709
91. Chang SC, Yang YC, Zhang ZF. Single nucleotide polymorphisms in genes of one-carbon metabolism pathway and bladder cancer survival. Cancer Epidemiol Biomarkers Prev. 2011;20:716-717
92. Babyshkina N, Malinovskaya E, Nazarenko M, Koval M, Gervas P, Potapova O. et al. The effect of folate-related SNPs on clinicopathological features, response to neoadjuvant treatment and survival in pre- and postmenopausal breast cancer patients. Gene. 2013;518:397-404
93. Zhao T, Xu Z, Gu D, Wu P, Huo X, Wei X. et al. The effects of genomic polymorphisms in one-carbon metabolism pathways on survival of gastric cancer patients received fluorouracil-based adjuvant therapy. Sci Rep. 2016;6:28019
94. de Jonge R, Hooijberg JH, van Zelst BD, Jansen G, van Zantwijk CH, Kaspers GJ. et al. Effect of polymorphisms in folate-related genes on in vitro methotrexate sensitivity in pediatric acute lymphoblastic leukemia. Blood. 2005;106:717-720
95. Stevens VL, McCullough ML, Pavluck AL, Talbot JT, Feigelson HS, Thun MJ. et al. Association of polymorphisms in one-carbon metabolism genes and postmenopausal breast cancer incidence. Cancer Epidemiol Biomarkers Prev. 2007;16:1140-1147
96. Stevens VL, Rodriguez C, Sun J, Talbot JT, Thun MJ, Calle EE. No association of single nucleotide polymorphisms in one-carbon metabolism genes with prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17:3612-3614
97. Babyshkina N. The effect of folate-related SNPs on therapeutic responses to neoadjuvant chemotherapy in breast cancer molecular subtypes. Mol Cancer Ther. 2011;10:C101-C101
98. Piskac-Collier AL, Monroy C, Lopez MS, Cortes A, Etzel CJ, Greisinger AJ. et al. Variants in folate pathway genes as modulators of genetic instability and lung cancer risk. Genes Chromosomes Cancer. 2011;50:1-12
99. DeRoo LA, Bolick SCE, Xu Z, Umbach DM, Shore D, Weinberg CR. et al. Global DNA methylation and one-carbon metabolism gene polymorphisms and the risk of breast cancer in the sister study. Carcinogenesis. 2014;35:333-338
100. Qu YY, Zhu SX, Zhang X, Zhao R, Gu CY, Chang K. et al. Functional variants of the 5-methyltetrahydrofolate-homocysteine methyltransferase gene significantly increase susceptibility to prostate cancer: Results from an ethnic Han Chinese population. Sci Rep. 2016;6:36264
101. Matsuo K, Hamajima N, Hirai T, Kato T, Inoue M, Takezaki T. et al. Methionine synthase reductase gene A66G polymorphism is associated with risk of colorectal cancer. Asian Pac J Cancer Prev. 2002;3:353-359
102. Gast A, Bermejo JL, Flohr T, Stanulla M, Burwinkel B, Schrappe M. et al. Folate metabolic gene polymorphisms and childhood acute lymphoblastic leukemia: a case-control study. Leukemia. 2007;21:320-325
103. Xu X, Gammon MD, Zhang H, Wetmur JG, Rao M, Teitelbaum SL. et al. Polymorphisms of one-carbon-metabolizing genes and risk of breast cancer in a population-based study. Carcinogenesis. 2007;28:1504-1509
104. Beetstra S, Suthers G, Dhillon V, Salisbury C, Turner J, Altree M. et al. Methionine-dependence phenotype in the de novo pathway in BRCA1 and BRCA2 mutation carriers with and without breast cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:2565-2571
105. Kotsopoulos J, Zhang WW, Zhang S, McCready D, Trudeau M, Zhang P. et al. Polymorphisms in folate metabolizing enzymes and transport proteins and the risk of breast cancer. Breast Cancer Res Treat. 2008;112:585-593
106. De Vogel S, Wouters KAD, Gottschalk RWH, Van Schooten FJ, De Goeij AFPM, De Bruïne AP. et al. Genetic variants of methyl metabolizing enzymes and epigenetic regulators: Associations with promoter CpG island hypermethylation in colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:3086-3096
107. Chen SQ, Dai WW, Xu SQ, Sun WW, Deng WY, Wu JW. et al. [The relationship of polymorphism of methionine synthase gene and plasma homocysteine level with intracranical aneurys]. Shi Yong Yi Xue Za Zhi. 2011;27:3522-3525
108. Mohammad NS, Yedluri R, Addepalli P, Gottumukkala SR, Digumarti RR, Kutala VK. Aberrations in one-carbon metabolism induce oxidative DNA damage in sporadic breast cancer. Mol Cell Biochem. 2011;349:159-167
109. Lopez-Cortes A, Jaramillo-Koupermann G, Munoz MJ, Cabrera A, Echeverria C, Rosales F. et al. Genetic polymorphisms in MTHFR (C677T, A1298C), MTR (A2756G) and MTRR (A66G) genes associated with pathological characteristics of prostate cancer in the Ecuadorian population. Am J Med Sci. 2013;346:447-454
110. Wu X, Zou T, Cao N, Ni J, Xu W, Tao Z. et al. Plasma homocysteine levels and genetic polymorphisms in folate metablism are associated with breast cancer risk in Chinese women. Hered Cancer Clin Pract. 2013;12:1198-1206
111. López-Cortés A, Echeverría C, Oña-Cisneros F, Sánchez ME, Herrera C, Cabrera-Andrade A. et al. Breast cancer risk associated with gene expression and genotype polymorphisms of the folate-metabolizing MTHFR gene: a case-control study in a high altitude Ecuadorian mestizo population. Tumor Biol. 2015;36:6451-6461
112. Hubner RA, Muir KR, Liu JF, Sellick GS, Logan RF, Grainge M. et al. Folate metabolism polymorphisms influence risk of colorectal adenoma recurrence. Cancer Epidemiol Biomarkers Prev. 2006;15:1607-1613
113. Kim YI. Folate and colorectal cancer: An evidence-based critical review. Mol Nutr Food Res. 2007;51:267-292
114. Yang Q, Bostick RM, Friedman J, Flanders WD. Serum folate and cancer mortality among US adults: findings from the Third National Health and Nutritional Examination Survey linked mortality file. Cancer Epidemiol Biomarkers Prev. 2009;18:1439-1447
115. Leclerc D, Wilson A, Dumas R, Gafuik C, Song D, Watkins D. et al. Cloning and mapping of a cDNA for methionine synthase reductase, a flavoprotein defective in patients with homocystinuria. Proc Natl Acad Sci U S A. 1998;95:3059-3064
116. Wilson A, Platt R, Wu Q, Leclerc D, Christensen B, Yang H. et al. A common variant in methionine synthase reductase combined with low cobalamin (vitamin B12) increases risk for spina bifida. Mol Genet Metab. 1999;67:317-323
117. Han D, Shen C, Meng X, Bai J, Chen F, Yu Y. et al. Methionine synthase reductase A66G polymorphism contributes to tumor susceptibility: evidence from 35 case-control studies. Mol Biol Rep. 2012;39:805-816
118. Wu PP, Tang RN, An L. A meta-analysis of MTRR A66G polymorphism and colorectal cancer susceptibility. J BUON. 2015;20:918-922
119. Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulosioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29:306-309
120. Colhoun HM, Mckeigue PM, Davey SG. Problems of reporting genetic associations with complex outcomes. Lancet. 2003;361:865-872
Author contact
![]() Corresponding authors: Shoumin Xi, The Key Laboratory of Pharmacology and Medical Molecular Biology, Medical College, Henan University of Science and Technology, No. 263 Kaiyuan Avenue, Luoyang 471023, Henan, China, Tel.: (+86-379) 64830346, Fax: (+86-379) 64830345, E-mail: xishouminedu.cn; or Jing He, Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, 9 Jinsui Road, Guangzhou 510623, Guangdong, China, Tel./Fax: (+86-20) 38076560, E-mail: hejing198374com.
Corresponding authors: Shoumin Xi, The Key Laboratory of Pharmacology and Medical Molecular Biology, Medical College, Henan University of Science and Technology, No. 263 Kaiyuan Avenue, Luoyang 471023, Henan, China, Tel.: (+86-379) 64830346, Fax: (+86-379) 64830345, E-mail: xishouminedu.cn; or Jing He, Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, 9 Jinsui Road, Guangzhou 510623, Guangdong, China, Tel./Fax: (+86-20) 38076560, E-mail: hejing198374com.

 Global reach, higher impact
Global reach, higher impact