3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(10):1317-1327. doi:10.7150/jca.15157 This issue Cite
Research Paper
Multi-target siRNA: Therapeutic Strategy for Hepatocellular Carcinoma
1. Department of Pathological Anatomy, Nantong University, Nantong, China;
2. Small RNA Technology and Application Institute, Nantong University, Nantong, China;
3. Biomics Biotechnologies Co., Ltd., Nantong, China.
Received 2016-2-1; Accepted 2016-5-18; Published 2016-6-25
Abstract
Multiple targets RNAi strategy is a preferred way to treat multigenic diseases, especially cancers. In the study, multi-target siRNAs were designed to inhibit NET-1, EMS1 and VEGF genes in hepatocellular carcinoma (HCC) cells. And multi-target siRNAs showed better silencing effects on NET-1, EMS1 and VEGF, compared with single target siRNA. Moreover, multi-target siRNA showed greater suppression effects on proliferation, migration, invasion, angiogenesis and induced apoptosis in HCC cells. The results suggested that multi-target siRNA might be a preferred strategy for cancer therapy and NET-1, EMS1 and VEGF could be effective targets for HCC treatments.
Keywords: multi-target siRNA, hepatocellular carcinoma (HCC), NET-1, EMS1, VEGF.
Introduction
RNA interference (RNAi) has become a popular approach recently, RNAi technology is applied for posttranscriptional gene silencing which triggered by small interfering RNA (siRNA) [1-3]. RNAi therapies have rapidly been advanced in clinical trials for the treatment of various human diseases, especially various human cancers [4-6], and the strategy of multiple gene-targeted siRNAs is considered to be a good way for controlling complex disease systems [7,8], such as hepatocellular carcinoma (HCC) [9].
HCC is the sixth most common malignancy tumor in the world and HCC has several complex and genetic alterations [10]. But the mechanisms of initiation and progression of HCC have not been made clear completely. Even though there are potential curative treatments, HCC can also easily become a poor prognosis tumor with the characteristics of high recurrence rate and rapid progress [11]. Therefore, development and identification of novel effective agents, such as siRNAs, that are able to suppress HCC effectively is imperative.
NET-1 gene is a member of the transmembrane 4 superfamily (TM4SF) [12], it is a tumor-related gene and found to be high expressed in many human tumor tissues [13-17]. NET-1 overexpression was also strong-related to pathological grading and clinical stages in HCC [18]. EMS1 is a cytoskeleton reorganization related gene, its coding protein is cortical actin-binding protein (cortactin) with binding ability to F-actin [19]. Studies found that cortactin could bind to Arp2/3, and influence cell motility, it also was considered has a relationship with cell migration [18]. Cortactin is a substrate of oncogene src, so its functions in tumors are studied deeply, and EMS1 was found high expressed in hepatocarcinoma, breast cancer and head and neck cancer, it had relationship with cancer invasion and transfer [20-23]. Vascular endothelial growth factor (VEGF) is a well-known gene in cancer therapy researches, angiogenesis inhibition is considered as an effective way for tumor treatment, thus VEGF is a good target for cancer therapy [24-26].
In the study, we intended to evaluate the effects of multiple targeting of NET-1, EMS1 and VEGF genes on biological behavior of HCC cell in-vitro.
Materials and Methods
Cell Lines and Cell Culture
Human embryonic kidney cells (HEK293), human HCC cells (MHCC97H, MHCC97L, HepG2, SMMC-7721) and human normal liver cells (LO2) were cultured in Dulbecco's Modified Eagle Medium (DMEM, Gibco) supplemented with 10 % fetal bovine serum (FBS, Gibco), 2 mM L-glutamine, 100 U/ml of penicillin and 100 μg/ml of streptomycin. These cell lines were purchased from the Institute of Cell Biology, Chinese Academy of Sciences, and cells were maintained at 37 °C in a humidified incubator with 5 % CO2.
Multi-target siRNA Design and siRNA Transfection
The sequence of NET-1 (Accssion No. NM 005727.3), EMS1 (Accssion No. NM 053056) and VEGF (Accssion No. NM 001287044.1) were obtained from National Center of Biotechnology Information (NCBI) GenBank (USA), and the single target siRNAs which targeting either NET-1, EMS1 or VEGF (NET-1_siR, EMS1_siR and VEGF_siR, 19 bp duplex with 2-nt 3'-overhangs) were designed and screened by our precious studies [27, 28]. And a siRNA which sequence has no homology with human genes was designed as negative control (NC_siR). All RNA oligonucleotides were chemically synthesized by Biomics Biotechnologies Co., Ltd. (Biomics Biotech, China). The oligonucleotide sequences were shown in Table 1. The cells were transfected in-vitro with siRNAs using Lipofectamine® 2000 transfection reagent (ThermoFisher, USA) according to the manufactures' instructions.
Multi-target siRNAs Design and Synthesis
The multi-target siRNA (mtg_siRs) constructs targeting NET-1, EMS1 and VEGF were designed according to the principle of Biomics Biotech (China) (sequences shown in Table 2). The long single stranded RNAs (the length was >21 nt) were synthesized by in vitro transcription mediated by T7 RNA polymerase. Briefly, T7 RNA polymerase promoter (5'-CACTAATACGACTCACTATAGGG-3') conjugated DNA oligos that complementary to the interest RNAs were chemically synthesized. The RNAs were synthesized by T7 RNA polymerase with the rNTPs at 37 °C for 5 h. Final RNA products were obtained post purification.
Sequences of the single target siRNAs.
| siRNAs | Sequences (5'-3') |
|---|---|
| NET-1_siR | Sense: CCACAAUGGCUGAGCACUUdTdT |
| Antisense: AAGUGCUCAGCCAUUGUGGdTdT | |
| EMS1_siR | Sense: GAACAAGACCGAAUGGAUAdTdT |
| Antisense: UAUCCAUUCGGUCUUGUUCdTdT | |
| VEGF_siR | Sense: GGAGUACCCUGAUGAGAUCdTdT |
| Antisense: GAUCUCAUCAGGGUACUCCdTdT | |
| NC_siR | Sense: UUCUCCGAACGUGUCACGUdTdT |
| Antisense: ACGUGACACGUUCGGAGAAdTdT |
Sequences of the Multi-target siRNAs (Mtg_siRNA)
| Mtg_siRNA | Targets | Sequences (5'-3') |
|---|---|---|
| NEV Str1 | NET-1 EMS1 VEGF | S:CCACAAUGGCUGAGCACUUGGAACAAGACCGAAUGGAUACGGAGUACCCUGAUGAGAUCdTdT |
| As1:AAGUGCUCAGCCAUUGUGGdTdT | ||
| As2:UAUCCAUUCGGUCUUGUUCdTdT | ||
| As3:GAUCUCAUCAGGGUACUCCdTdT | ||
| NEV Str2 | NET-1 EMS1 VEGF | S:CCACAAUGGCUGAGCACUUGGAACAAGACCGAAUGGAUACGGAGUACCCUGAUGAGAUCdTdT |
| As1:GAUCUCAUCAGGGUACUCCdTdT | ||
| As2:UAUCCAUUCGGUCUUGUUCUUCAAGAGAAAGUGCUCAGCCAUUGUGGdTdT | ||
| NEV Str3 | NET-1 EMS1 VEGF | S:CCACAAUGGCUGAGCACUUGGAACAAGACCGAAUGGAUACGGAGUACCCUGAUGAGAUCdTdT |
| As1:AAGUGCUCAGCCAUUGUGGdTdT | ||
| As2:GAUCUCAUCAGGGUACUCCUUCAAGAGAUAUCCAUUCGGUCUUGUUCdTdT |
S, Sense Strand; As, Antisense Strand.
Dual Luciferase Reporter Assay
The siRNA validation vector siQuant (kindly provided by Dr. Quan Du of Peking University) [29] was used for detection of the gene silencing activity of mtg_siRs. When the siRNA validation vectors (siQNET-1, siQEMS1 and siQVEGF) were constructed, the activities of mtg_siRs were analyzed by dual luciferase reporter (DLR) assay according to the protocol of Du Q et al. [30]. Briefly, the DNA oligonucleotides were designed (Table 3), synthesized (Biomics Biotech, China) and annealed to DNA duplex. Then they were ligated to construct siRNA validation vectors containing siRNA target site of NET-1, EMS1 or VEGF, and all constructs were verified by sequencing. HEK293 cells were grown and seeded into 24-well plates at 1×105 cells/well for 24 h before transfection. The validation vector was co-transfected with pRL-TK (Promega, USA) with or without siRNAs. The cells were harvested after 24 h, according to the DLR assay manual (Promega, USA). And the activities of luciferase were measured on a Luminometer (Promega, USA). The firefly luciferase signal was normalized to the renilla luciferase signal for each individual well, and the knockdown efficacy of each siRNA was evaluated by relative luciferase activity unit (RLU). All experiments were performed in triplicates and repeated three times.
Sequences of the siQuant inserts.
| siQuant Reporter | Sequences (5'-3') |
|---|---|
| siQNET-1 | S: GATCTCACCACAATGGCTGAGCACTTCCGGGCC |
| As: CGGAAGTGCTCAGCCATTGTGGTGA | |
| siQEMS1 | S: ATCTCAAGAACAAGACCGAATGGATACGGGCC |
| As: CGTATCCATTCGGTCTTGTTCTTGA | |
| siQVEGF | S: GATCTCAAAGGAGTACCCTGATGAGATCGGGCC |
| As: CGATCTCATCAGGGTACTCCTTTGA |
S, Sense Strand; As, Antisense Strand.
Real-time Quantity PCR (RT-qPCR)
Total RNA of cells were extracted by RISOTM RNA extraction reagent (Biomics Biotech, China) and then performed to a 25 μL PCR reaction: 12.5 μL of 2×Master Mix (Biomics Biotech, China), 0.5 μL of each forward and reverse primers (10 μM each), 0.5 μL of 50×SYBR Green I and 4 μL total RNA, then subjected to reverse transcription for 30 min at 42 °C and initially denatured at 95 °C for 5 min, and then to 45 cycles of amplification with the condition of: 95 °C denature for 20 s, 55 °C annealing for 30 s, and 72 °C extension for 30 s. Human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as an internal control. All primers sequences were shown in Table 4. The experiment was performed in triplicate. The results were analyzed by 2-ΔΔCt method [31].
Sequences of RT-qPCR primers.
| Gene Name | Sequence (5'-3') | |
|---|---|---|
| NET-1 | Forward | CTTCATCCTCCTCCTCATCTTC |
| Reverse | CAGGCACTACCAGCAACG | |
| EMS1 | Forward | AGAGGCTGTCTATGAAAG |
| Reverse | GATGTTGGTGATGATGTC | |
| VEGF | Forward | GACATCTTCCAGGAGTACC |
| Reverse | TGCTGTAGGAAGCTCATCTC | |
| Cyclin D1 | Forward | ATCTACACCGACAACTCCATC |
| Reverse | TGTTCTCCTCCGCCTCTG | |
| bFGF | Forward | AGCAGCATCTGTAAGGTTCTTC |
| Reverse | TGAAACATTGGGAGGGAAAC | |
| Fascin | Forward | GCTGGAGTTCAACGATGG |
| Reverse | ACCTTGTTATAGTCGCAGAAC | |
| OAS1 | Forward | GTGAGCTCCTGGATTCTGCT |
| Reverse | TGTTCCAATGTAACCATATTTCTGA | |
| IFIT1 | Forward | AATAGACTGTGAGGAAGGATGG |
| Reverse | TCCAGGCGATAGGCAGAG | |
| GAPDH | Forward | GAAGGTGAAGGTCGGAGTC |
| Reverse | GAAGATGGTGATGGGATTTC |
Western Blot
1×106 cells/well were plated in a 6-well plate and grown for 24 h until about 70% confluence. After cells were treated by siRNAs for 48 h, they were harvested and lysed in RIPA buffer (Pierce, USA). Total protein was separated by polyacrylamide gel electrophoresis (PAGE) and electroblotted onto polyvinylidine difluoride filter (PVDF) membranes (Millipore, USA), followed by blocking with 5% skim milk in TBST (20 mM Tris, 150 mM NaCl, 0.05% Tween-20, pH7.5) buffer for 2 h at room temperature, then incubated with rabbit-anti-human NET-1 antibody (1:500 dilution, Abgent, USA), rabbit-anti-human EMS1 (1:500 dilution, Abcam, USA) and rabbit-anti-human VEGF antibody (1:500 dilution, Abcam, USA), mouse-anti-human β-actin antibody (1:500 dilution, Abcam, USA) as internal control. Then membranes were washed in TBST and incubated with a goat anti-rabbit or goat anti-mouse HRP-conjugated secondary antibody (1:1000 dilution, Jackson, USA) at room temperature for 2 h. At last, the specific proteins were detected with ECL chemiluminescence reagent (Beyotime, China); the membranes were exposed to films (Kodak, USA).
Immunocytochemistry (ICC)
7.5×104 cells were cultured on a small round slide in a 24-well plate, when cells grown to about 50% confluence, they were treated as described above. Then cells were fixed with 4% paraformaldehyde for 30 min and permeabilized with 0.5 % Triton X-100 for 10 min, after being washed by PBS, blocked with 1% bovine serum albumin (BSA) for 30 min. Subsequently, cells were incubated at 4 °C overnight with antibodies as used in Western blot, and then with TRITC-conjugated goat-anti-rabbit IgG (for NET-1 or VEGF) and FITC-conjugated goat-anti-rabbit IgG (for EMS1) (1:50 dilution, Boster, China) for 2 h at room temperature. Hoechst 33258 (10 mg/mL) (ThermoFhisher, USA) was used for cell nuclei staining, after washed by PBS and mounted by Antifade Mounting Medium (Beyotime, China), cells were observed under immunofluorescence microscopy.
Cell Viability Assay
The viability of cells was measured by MTT assay. 5×103 cells/well were plated into a 96-well plate for 24 h, then treated by siRNAs for 0, 24, 48, 72 and 96 h, 10 μL MTT solution (Beyotime, China) were added to each well of 96-well plate containing 100 μL DMEM and incubated at 37 °C for 4 h, following 150 μL DMSO per well were added and continuing incubated at 37 °C for 10 min. The optical density (OD) was measured at 490 nm using a Microplate Reader (BioRad, USA).
Cell Invasion Assay
2×104 cells/well were seeded into a 24-well plate for 24 h and treated as described above for 48 h, then cells were suspended in DMEM at the density of 1×106 cells/mL. Briefly, transwell chambers (Corning, USA) were treated with DMEM for 1 h before treatments; the upper and lower chambers were separated by an 8 μm pore polycarbonate membrane which was coated with 50 μL of 0.5 mg/ml matrigel (BD, USA). 100 μL cell suspension was added into each upper chamber with 600 μL DMEM containing 10% FBS or conditioned medium which were the cell supernatant with siRNAs to perform post-transfection for 48 h. The cells on the top surface of the membrane were carefully removed after 24 h post treatments. Cells on the transwell chambers were fixed for 30 s in 10% formaldehyde, and then stained by 0.5% crystal violet. After being washed by PBS, the cells on the top surface of the membrane were carefully removed again. The cells on the bottom surface of the membrane were counted in 3-5 random fields under microscope (magnification 100×).
Cell Apoptosis Assay
Annexin V-FITC/PI double staining and flow cytometry (FCM) analysis method was used to determine the cell apoptosis. Briefly, 1×106 cells/well in a 6-well plate treated with siRNAs for 48 h were harvested and washed in PBS, then re-suspended in binding buffer, followed by incubation with Annexin V-FITC conjugate and PI for 15 min, then detected by FCM analysis (BD Biosciences, USA).
Tube Formation Assay
HUVECs in-vitro angiogenesis model was used to evaluate inhibition effect. Briefly, 1×105 HUVECs were re-suspended in conditioned medium (which was the supernatant of cells treated with siRNAs for 48 h), then seeded in a 48-well plate which were coated by matrigel (BD, USA), and cultured in at 37 °C/5% CO2 incubator for 24 h, the numbers of branching points were counted under microscope (magnification 40×).
Cell Migration Assay
Wound-healing assay was used to detect the migration inhibition effect. Briefly, 3×105 cells/well were plated and treated as described above in a 12-well plate. Wound was made through confluent monolayer cells with a pipette tip post-transfected for 48 h and cells were washed with DMEM twice. Cell photos were taken at 0, 24, 48, and 72 h to monitor cell migration.
Statistical Analysis
All experiments were performed independently three times, the results were shown as mean ± standard deviation (SD), and statistical analyses were performed using SPSS19.0 software. The differences between two groups were compared using Student's t-test and multiple comparisons using one-way ANOVA analysis to assess statistical significance. All P values were based on a two-sided statistical analysis and P<0.05 was considered to indicate statistical significance.
Results
Multi-target siRNAs were designed and synthesized by chemical and in vitro transcription method
The multi-target siRNAs (mtg_siRs) targeting NET-1, EMS1 and VEGF were designed according to different structures (Fig. 1). The short single stranded RNAs and long single stranded RNAs (the length was >21 nt) of mtg_siRs were synthesized by chemical and in vitro transcription method separately, and mtg_siRs were obtained by annealing long RNA and corresponding short RNAs (Table 2, Fig. 1).
mtg_siRs were designed and synthesized by chemical and in vitro transcription method. (A) The designed different mtg_siRs structures, bold lines are sense strands and the thin lines are antisense strands, (B) The synthesitcal mtg_siRs detected by 15% polyacrylamide gelelectrophoresis (PAGE).
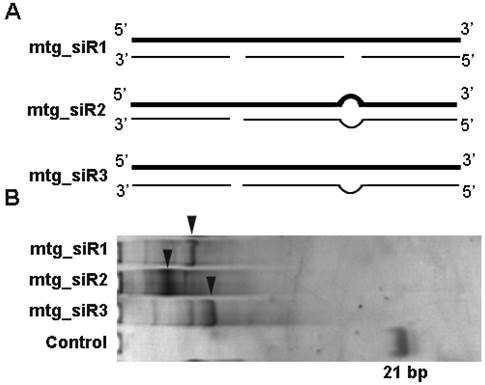
The designed mtg_siRs validation vectors of siQuant method. (A) The designed siRNA target (NET-1, EMS1 and VEGF) siQuant vectors, (B) The sequencing results of different siRNA validation vectors (siQNET-1, siQEMS1 and siQVEGF).
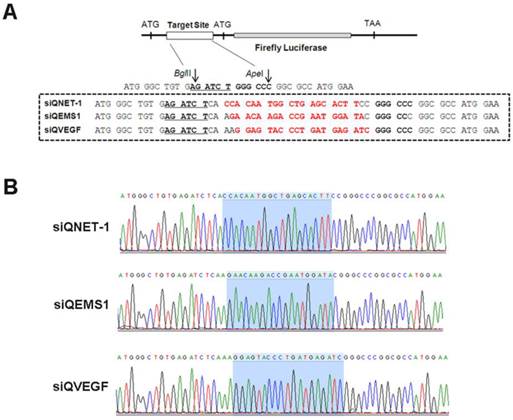
The activities of different multi-target siRNAs detected by DLR assay
The siRNA validation vectors and mtg_siRs were co-transfected into HEK293 cells, and the activities detected by DLR system (Promega, USA). As shown in Fig. 2 and 3, the siRNA validation vectors (siQNET-1, siQEMS1 and siQVEGF) were constructed successfully and the activities of mtg_siRs were indicated by RLU values with different treatments. Compared with control (CTL) and NC_siR, NET-1 inhibition rates of mtg_siR1, mtg_siR2 and mtg_siR3 were 60.95%, 57.49% and 81.13% (P<0.05), EMS1 inhibition rates of mtg_siR1, mtg_siR2 and mtg_siR3 were 70.61%, 62.32% and 64.44% (P<0.05), VEGF inhibition rates of mtg_siR1, mtg_siR2 and mtg_siR3 were 82.91%, 86.73% and 77.76% (P<0.05). The specific gene knockdown could be interfered by interferon (IFN) response induced by long stranded dsRNA [32], thus the expression of OAS1 and IFIT1 genes were used to monitor the IFN response induced by mtg_siRs. 10 ng/mL Poly(I:C) [33] transfection served as positive control. Results showed no obvious IFN response induced by mtg_siRs, compared with positive or negative control (Fig. 3D).
Gene inhibition effects in HCC cell by multi-target siRNAs
The result of RT-qPCR showed that, compared with normal hepatocyte LO2, the mRNA level of NET-1, EMS1 and VEGF were all high expressed in HCC cells (MHCC97H, MHCC97L, HepG2 and SMMC-7721) (P<0.05), especially in SMMC-7721, NET-1 was 7.31-fold, EMS1 was 4.61-fold and VEGF was 7.67-fold higher than in LO2 cell (P<0.05, Fig. 4A), SMMC-7721 cell was chosen to further study.
As shown in Fig. 4B-D, compared to untreated group, NET-1 mRNA was inhibited by NET-1_siR, mtg_siR1, mtg_siR2 and mtg_siR3 up to 27%, 31%, 25% and 33% (P<0.05). EMS1 was inhibited by EMS1_siR, mtg_siR1, mtg_siR2 and mtg_siR3 up to 28%, 14%, 37% and 24% (P<0.05). VEGF was inhibited by VEGF_siR, mtg_siR1, mtg_siR2 and mtg_siR3 up to 29%, 18%, 17% and 19% (P<0.05), and there was no difference among mtg_siR1, mtg_siR2 and mtg_siR3 and there was no difference between NC_siR and untreated group (P>0.05).
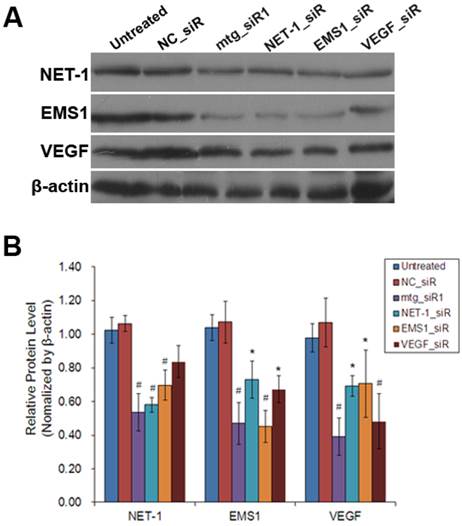
As shown in Fig. 5, the results of Western blot showed that, compared to untreated group, NET-1 protein level was inhibited by mtg_siR1, NET-1_siR, EMS1_siR and VEGF_siR up to 54%, 58%, 70% and 84% (P<0.05). EMS1 was inhibited by mtg_siR1, NET-1_siR, EMS1_siR and VEGF_siR up to 47%, 73%, 45% and 67% (P<0.05). VEGF was inhibited by mtg_siR1, NET-1_siR, EMS1_siR and VEGF_siR up to 39%, 69%, 71% and 48% (P<0.05), and there was no difference between NC_siR and untreated group (P>0.05).
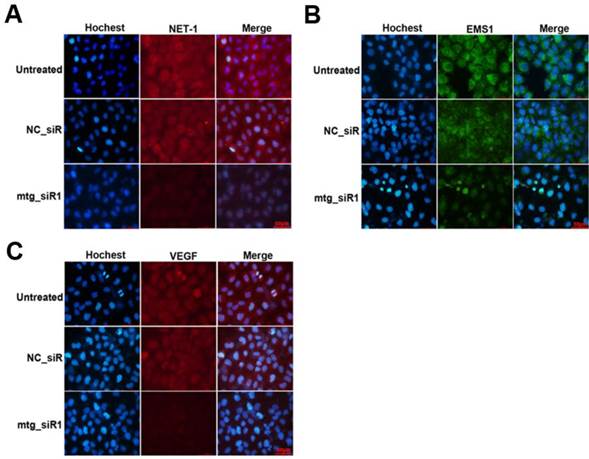
The inhibition effects on the protein level of target genes were further determined by ICC method. As show in Fig. 6, the protein of NET and VEGF were labeled by TRITC (red), and EMS1 was labeled by FITC (green), the cell nuclei stained by Hoechst (blue). The results showed that the expression of NET-1, EMS1 and VEGF were inhibited by mtg_siR1, compared to NC_siR and untreated group.
The inhibition activities of mtg_siRs validated by DLR assay. (A) siQNET-1 inhibited by mtg_siRs, (B) siQEMS1 inhibited by mtg_siRs, (C) siQVEGF inhibited by mtg_siRs, *P<0.05, (D) No interferon response induced by mtg_siRs.
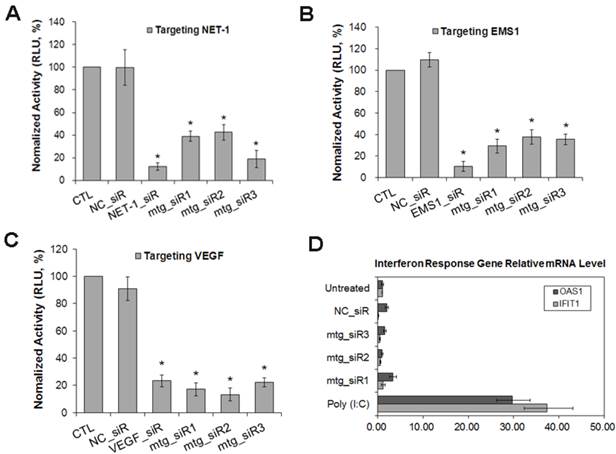
The mRNA expression level of NET-1, EMS1 and VEGF in HCC cells detected by RT-qPCR. (A) The mRNA expression level of NET-1, EMS1 and VEGF in different HCC cells, *P<0.05 vs LO2, (B) The expression of NET-1 inhibited by mtg_siRs in mRNA level, (C) The expression of EMS1 inhibited by mtg_siRs in mRNA level, (D) The expression of VEGF inhibited by mtg_siRs in mRNA level. *P<0.05 vs untreated group.
The inhibition of NET-1, EMS1 and VEGF in protein level. (A) The inhibition of NET-1, EMS1 and VEGF in protein level measured by Western blot, (B) The inhibition rate of different treatments, *P<0.05 vs untreated group.
Inhibition effects of target gene by multi-target siRNA in HCC cell measured by ICC. (A) The expression of NET-1 inhibited by mtg_siR1, (B) The expression of EMS1 inhibited by mtg_siR1, (C) The expression of VEGF inhibited by mtg_siR1.
Proliferation and apoptosis relative genes regulated by multi-target siRNA
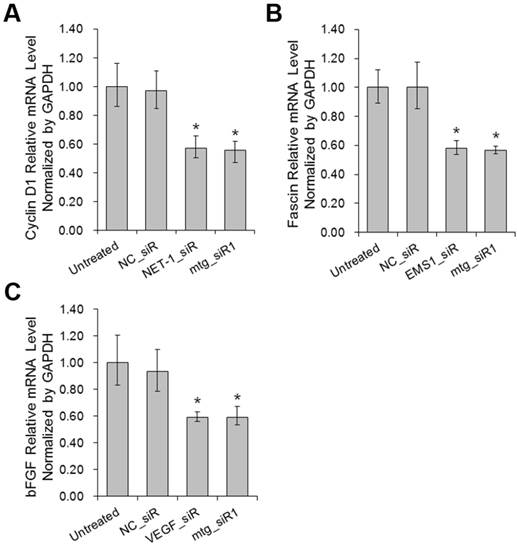
The mRNA expression levels of Cyclin D1, Fascin and bFGF were determined by RT-qPCR post siRNA treatments. The expression of Cyclin D1, Fascin and bFGF were all down-regulated on different level (P<0.05) by mtg_siR1 and corresponding single target siRNA (Fig. 7), compared with NC_siR and untreated group.
Inhibition effect of cell viability by multi-target siRNA
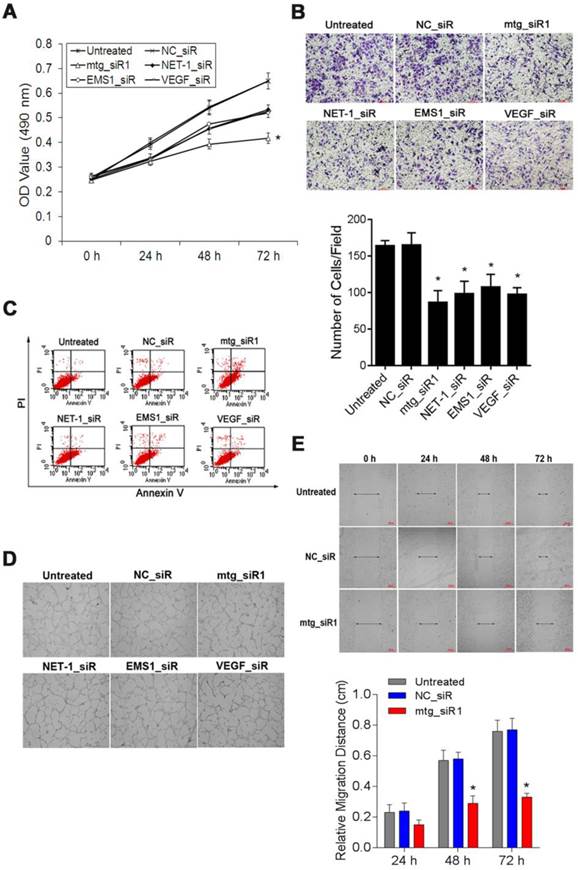
The viability of SMMC-7721 cells was measured by MTT assay. The absorbance values (490 nm) of the cells at 48, 72 and 96 h post-transfection with mtg_siR1, NET-1_siR, EMS1_siR and VEGF_siR were significantly lower than those of NC_siR treated cells and untreated cells respectively. There was no significant difference between the growth of cells treated with NET-1_siR, EMS1_siR and VEGF_siR, and the cell viability in mtg_siR1 treated group showed a significant decrease (P<0.05, Fig. 8A).
Inhibition effect of cell invasion by multi-target siRNA
The cell invasion abilities with different treatments were detected by transwell assay, as Fig. 8B showed that the invasion of the cells treated with mtg_siR1, NET-1_siR, EMS1_siR and VEGF_siR were inhibited significantly post 48 h transfection (P<0.05). There was no significant difference of inhibition abilities between NC_siR and untreated cells (P>0.05), and the number of invasion cells in mtg_siR1 treated group was significantly decreased than that of NET-1_siR, EMS1_siR and VEGF_siR (P<0.05).
Cell apoptosis induced by multi-target siRNA
Annexin V-FITC/PI double staining and FCM analysis were used to detect the ability of mtg_siR on inducing SMMC-7721 cell apoptosis. As shown in Fig. 8C, treatments with mtg_siR1 (28.93±1.93), NET-1_siR (15.46±1.47%), EMS1_siR (10.64±2.91), and VEGF_siR (15.82±1.40) resulted in a significant increase of apoptosis compared with that of NC_siR treated cells (3.08±0.97%) and untreated cells (2.40±0.54%) (P<0.05), and the apoptosis rate of mtg_siR1 treated group were higher than that of NET-1_siR, EMS1_siR and VEGF_siR (P<0.05).
The affection of proliferation and apoptosis relative genes when down-regulation of NET-1, EMS1 and VEGF detected by RT-qPCR. (A) The mRNA level of proliferation gene Cyclin D1, (B) The mRNA level of motility gene Fascin, (C) The mRNA level of fibroblast growth factor bFGF, *P<0.05 vs untreated group.
The suppression effects on cell viability, invasion, angiogenesis, migration and induced apoptosis by mtg_siR. (A) Cell growth curve of all groups at 0 h, 24 h, 48 h, 72 h, (B) Representative field photograph of transwell chamber assay under single-target siRNA and mtg_siR treatment after 48 h, (C) Cell apoptosis was measured by FCM analysis after AnnexinV/PI staining, (D) The tube formation photographs of each treatment in HUVECs angiogenesis model, (E) The ability of cell migration at 0 h, 24 h, 48 h, 72 h.
Influence effect of multi-target siRNA on tube formation
The tube formation was evaluated in HUVEC angiogenesis model, HUVECs stimulated by the conditioned medium derived from SMMC-7721 cells transfected with mtg_siR1, NET-1_siR, EMS1_siR and VEGF_siR. As illustrated in Fig. 8D, HUVECs treated with the conditioned medium of mtg_siR1, NET-1_siR, EMS1_siR and VEGF_siR were inhibited to form extensive and enclosed tube networks as compared with the untreated ones. The numbers of branching points were counted that mtg_siR1 (11.00±2.00), NET-1_siR (27.67±1.53), EMS1_siR (27.33±4.51), and VEGF_siR (15.33±2.08) less than NC_siR (69.33±3.06) and untreated cells (72.00±3.61) significantly (P<0.05).
Inhibition effect of cell migration by multi-target siRNA
The migration ability of SMMC-7721 cells inhibited by mtg_siR was measured by wound-healing assay. The scratch caused in groups of untreated and NC_siR nearly closed completely after 72 h, but the cells in treatment with NET-1_siR, EMS1_siR and VEGF_siR were not able to move toward the center of the wound. Compared to the NC_siR treated and untreated cells, the abilities of wound healing in NET-1_siR, EMS1_siR and VEGF_siR treated cells were decreased, and the migration distance in mtg_siR1 treated cells was significantly shorter than that of NET-1_siR, EMS1_siR and VEGF_siR (Fig. 8E).
Discussion
Multi-targeting therapies for cancers have been considered to be an effective approach, and RNAi strategy is a better method for multiple genes suppression, and there are many siRNA drugs in clinical trials which two or more gene inhibition simultaneously [8]. On the other hand, structural modified siRNAs had been widely used in RNAi therapeutics development, especially long double-stranded RNAs (dsRNA) were designed for carrying two or more siRNA sequences which targeting two or more parts in one mRNA or two or more mRNAs [34-36]. Current studies showed that, once long dsRNAs get into mammalian cells, they can be used as Dicer's substrates for siRNAs processing, then the corresponding genes could be inhibited effectively by siRNAs [1,2,8].
In our study, we designed a series of multi-target siRNA structures for triple cancer genes inhibition in hepatocellular carcinoma. We chose NET-1, EMS1 and VEGF as three target genes. NET-1 was reported as a cancer cell proliferation related gene [37], EMS1 was a cancer cell motility and transfer gene [28] and VEGF was angiogenesis related gene in tumor tissues [25]. In our previous studies, we designed and screened better siRNAs that targeting NET-1, EMS1 or VEGF gene respectively [27, 28]. Here, the multi-target siRNAs were designed for carrying on three different siRNA sequences which targeting NET-1, EMS1 and VEGF genes (Table 2, Fig. 1). In detail, three different long dsRNA structures containing three functional siRNA targets, but long RNA chemical synthesis method currently cannot meet our purpose, so the long RNAs were synthesized by T7 polymerase-mediated in vitro transcription (as shown in Fig. 1, know-how technology of Biomics Biotechnologies Co., Ltd).
To validate the functions of designed multi-target siRNA structures accurately and quickly, we used a dual luciferase reporter (DLR) assay based on siQuant method [29]. The result showed that three designed structures (mtg_siR1, mtg_siR2 and mtg_siR3) were all have gene silencing efficiencies (Fig. 3). Then, we screened a HCC cell line (SMMC-7721) with high expressed NET-1, EMS1 and VEGF simultaneously for in-vitro functional study (Fig. 4). The mRNA and protein levels of NET-1, EMS1 and/or VEGF were suppressed by mtg_siR, NET-1_siR, EMS1_siR and VEGF_siR respectively (Fig. 4-6). And the proliferation gene (Cyclin D1), motility gene (Fascin) and fibroblast growth factor (bFGF) were determined to confirm the inhibition effects of the three genes (Fig. 7).
Furthermore, we demonstrated that the proliferation, migration, invasion and induced apoptosis of HCC cells were inhibited by multi-target siRNA (mtg_siR1) effectively, and the angiogenesis was also suppressed by it (Fig. 8).
The results of the in-vitro study were demonstrated that the designed multiple targeting siRNA (mtg_siR1) construct had RNAi activities on knockdown triple genes simultaneously. Moreover, NET-1, EMS1 and VEGF gene silencing simultaneously triggered by siRNA was much better than single target siRNA in the inhibition of either NET-1, EMS1 or VEGF gene. And, co-silencing of NET-1, EMS1 and VEGF could suppress the proliferation, migration, invasion and angiogenesis, while inducing apoptosis of HCC cells in-vitro.
The results showed that, multi-target siRNA might be a preferred strategy for multigenic disease therapy for cancers. Also, the result suggested that NET-1, EMS1 and VEGF would be effective target genes for HCC therapy.
Acknowledgements
This study was supported by the foundation of the production-study-research prospective joint research programs of Jiangsu Province, China (No. BY 2013042-06), and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, and from the Science Foundation of Nantong City, Jiangsu Province, China (nos. BK 2014001 and HS 2014004).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Fire A, Xu S, Montgomery MK. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806-811
2. Hammond SM, Bernstein E, Beach D. et al. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293-296
3. Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188-200
4. Takeshita F, Ochiya T. Therapeutic potential of RNA interference against cancer. Cancer Sci. 2006;97:689-696
5. Aigner A. Applications of RNA interference: current state and prospects for siRNA-based strategies in vivo. Appl Microbiol Biotechnol. 2007;76:9-21
6. Davidson BL, MCCRAY PB. Current prospects for RNA interference-based therapies. Nat Rev Genet. 2011;12:329-340
7. Zimmermann GR, Leha RJ, Keith CT. Multi-target therapeutics: when the whole is greater than the sum of the parts. Drug Discov Today. 2007;12:34-42
8. Li T, Wu M, Zhu YY. et al. Development of RNA interference-based therapeutics and application of multi-target small interfering RNAs. Nucleic Acid Ther. 2014;24:302-312
9. Tabernero J, Shapiro GI, LoRusso PM. et al. First-in-humans trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement. Cancer Discov. 2013;3:406-417
10. Forner A, Gilabert M, Bruix J. et al. Treatment of intermediate-stage hepatocellular carcinoma. Nat Rev Clin Oncol. 2014;11:525-535
11. Bruix J, Gores GJ, Mazzaferro V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut. 2014;63:844-855
12. Yauch RL, Hemler ME. Specific interactions among transmembrane 4 superfamily (TM4SF) proteins and phosphoinositide 4-kinase. Biochem J. 2000;35:629-637
13. Chen L, Wang Z, Zhan X. et al. Association of NET-1 gene expression with human hepatocellular carcinoma. Int J Surg Pathol. 2007;15:346-353
14. Chen L, Yuan D, Wang GL. et al. Clinicopathological significance of overexpression of TSPAN1, Ki67 and CD34 in gastric carcinoma. Tumori. 2008;94:529-536
15. Chen L, Zhu YY, Zhang XJ. et al. TSPAN1 protein expression: a significant prognostic indicator for patients with colorectal adenocarcinoma. World J Gastroenterol. 2009;15:2270-2276
16. Scholz CJ, Kurzeder C, Koretz K. et al. Tspan-1 is a tetraspanin preferentially expressed by mucinous and endometrioid subtypes of human ovarian carcinomas. Cancer Lett. 2009;275:198-203
17. Zhang J, Wang J, Chen L. et al. Expression and function of NET-1 in human skin squamous cell carcinoma. Arch Dermatol Res. 2014;306:385-397
18. van Rossum AG, Schuuring-Scholtes E, van Buuren-van Seggelen V. et al. Comparative genome analysis of cortactin and HS1: the significance of the Factin binding repeat domain. BMC Genomics. 2005;6:15
19. Chuma M, Sakamoto M, Yasuda J. et al. Overexpression of cortactin is involved in motility and metastasis of hepatocellular carcinoma. J Hepatol. 2004;41:629-636
20. Chen L, Wang Z, Zhu J. et al. Roles of cortactin, an actin polymerization mediator, in cell endocytosis. Acta Biochim Biophys Sin (Shanghai). 2006;38:95-103
21. Zhu J, Zhou K, Hao JJ. et al. Regulation of Cortactin/Dynamin interaction by actin polymerization during the fission of clathrin-coated pits. J Cell Sci. 2005;118:807-817
22. Hao JJ, Zhu J, Zhou K. et al. The coiled-coil domain is required for HS1 to bind to F-actin and activate Arp2/3 complex. J Biol Chem. 2005;280:37988-37994
23. Lin J, Liu J, Wang Y. et al. Differential regulation of Cortactin and WASP mediated actin polymerization by missing in metastasis (MIM) protein. Oncogene. 2005;24:2059-2066
24. Yasuda S, Arii S, Mori A. et al. Hexokinase II and VEGF expression in liver tumors: correlation with hypoxia-inducible factor 1 alpha and its significance. J Hepatol. 2004;40:117-123
25. Rapisarda A, Melillo G. Role of the VEGF/VEGFR axis in cancer biology and therapy. Adv Cancer Res. 2012;114:237-267
26. Sitohy B, Nagy JA, Dvorak HF. Anti-VEGF/VEGFR therapy for cancer: reassessing the target. Cancer Res. 2012;72:1909-1914
27. Wu YY, Chen L, Wang GL. et al. Inhibition of hepatocellular carcinoma growth and angiogenesis by dual silencing of NET-1 and VEGF. J Mol Histol. 2013;44:433-445
28. Zhou J, Chen L, Zhang Y. et al. Synergistic effect of EMS1-shRNA and sorafenib on proliferation, migration, invasion and endocytosis of SMMC-7721. J Mol Histol. 2014;45:205-216
29. Du Q, Thonberg H, Zhang HY. et al. Validating siRNA using a reporter made from synthetic DNA oligonucleotides. Biochem Biophys Res Commun. 2004;325:243-249
30. Du Q, Thonberg H, Wang J. et al. A systematic analysis of the silencing effects of an active siRNA at all single-nucleotide mismatched target sites. Nucleic Acids Res. 2005;33:1671-1677
31. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408
32. Elbashir SM, Harborth J, Lendeckel W. et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494-498
33. Palchetti S, Starace D, De Cesaris P. et al. Transfected poly(I:C) activates different dsRNA receptors, leading to apoptosis or immunoadjuvant response in androgen-independent prostate cancer cells. J Biol Chem. 2015;290:5470-83
34. Shin D, Lee H, Kim SI. et al. Optimization of linear double-stranded RNA for the production of multiple siRNAs targeting hepatitis C virus. RNA. 2009;15:898-910
35. Chang CI, Kang HS, Ban C. et al. Dual-target gene silencing by using long, synthetic siRNA duplexes without triggering antiviral responses. Mol Cells. 2009;27:689-695
36. Rice RR, Muirhead AN, Harrison BT. et al. Simple, robust strategies for generating DNA-directed RNA interference constructs. Methods Enzymol. 2005;392:405-419
37. Chen L, Li X, Wang GL. et al. Clinicopathological significance of overexpression of TSPAN1, Ki67 and CD34 in gastric carcinoma. Tumori. 2008;94:529-536
Author contact
![]() Corresponding authors: Li Chen, Professor. Department of Pathological Anatomy, Nantong University, 19 Qixiu Road, Nantong 226001, Jiangsu, China. Email: bl1edu.cn. York Yuanyuan Zhu, Ph.D. Small RNA Technology and Application Institute, Nantong University, 76 Changxing Road, E&T Development Area, Nantong 226016, China. Email: yzhucom. Tel: +86-513-85051729; Fax: +86-513-85051729.
Corresponding authors: Li Chen, Professor. Department of Pathological Anatomy, Nantong University, 19 Qixiu Road, Nantong 226001, Jiangsu, China. Email: bl1edu.cn. York Yuanyuan Zhu, Ph.D. Small RNA Technology and Application Institute, Nantong University, 76 Changxing Road, E&T Development Area, Nantong 226016, China. Email: yzhucom. Tel: +86-513-85051729; Fax: +86-513-85051729.

 Global reach, higher impact
Global reach, higher impact