3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(5):490-499. doi:10.7150/jca.12351 This issue Cite
Research Paper
Evaluation of Plasma miR-21 and miR-152 as Diagnostic Biomarkers for Common Types of Human Cancers
1. Rush University Cancer Center, Chicago, IL 60612;
2. Department of Pathology, Rush University Medical Center, Chicago, IL 60612;
3. Department of Cardiothoracic Surgery, Rush University Medical Center, Chicago, IL 60612;
4. Department of Biochemistry, Rush University Medical Center, Chicago, IL 60612;
5. Department of Internal Medicine, Rush University Medical Center, Chicago, IL 60612;
6. Division of Urology, Department of Surgery, University of Arizona Cancer Center, Tucson, AZ 85724;
7. Department of Psychiatry, School of Medicine, the Johns Hopkins University, Baltimore, MD 21287.
8. Shenzhen Baoan District Shajing Hospital, Shenzhen, Guangdong 518104, China.
9. Department of Clinical Laboratory, The Fourth Hospital Affiliated to Guangxi Medical University, Liuzhou, Guangxi 545005, China.
*These authors had equal contribution to the presented work.
Received 2015-4-8; Accepted 2015-6-11; Published 2016-2-5
Abstract
Stable blood based miRNA species have allowed for the differentiation of patients with various types of cancer. Therefore, specific blood-based miRNA might be considered as a methodology which could be informative of the presence of cancer potentially from multiple distinct organ sites. Recently, miR-21 has been identified as an “oncomir” in various tumors while miR-152 as a tumor suppressor. In this study, we investigated whether circulating miR-21 and miR-152 can be used for early detection of lung cancer (LuCa), colorectal carcinoma (CRC), breast cancer (BrCa) and prostate cancer (PCa), with distinguishing cancer from various benign lesions on these organ sites. We measured the two miRNA levels by using real-time RT-PCR in plasma samples from a total of 204 cancer patients, 159 various benign lesions, and 228 normal subjects. We observed significantly elevated expression of miR-21 and miR-152 in LuCa, CRC, and BrCa when compared with normal controls. We also found upregulation of plasma miR-21 and miR-152 levels in patients with benign lesions of lung and breast, as compared to normal controls, respectively. No significant expression variation of the two miRNAs was observed in PCa or prostatic benign lesions as compared to healthy controls. Receiver operating characteristic (ROC) analyses revealed that miR-21 and/or miR-152 can discriminate LuCa, CRC and BrCa from normal controls. Our results suggest that plasma miR-21 and miR-152 may serve as non-specific noninvasive biomarkers for early screening of LuCa, CRC, and BrCa, but not PCa.
Keywords: miRNA, cancer
Introduction
Cancer is a severe health problem threatening human life in the United States and many other parts of the world. Among men in the United States, the three most common cancers are prostate, lung and bronchus, and colorectal cancers, which account for about 50% of all newly diagnosed cancers with prostate cancer (PCa) for 28% of incident cases alone1. The three most commonly diagnosed types of cancer among women are breast, lung and bronchus, and colorectal, accounting for 51% of estimated cancer cases with breast cancer (BrCa) alone for 29% of all new cancer cases1. Early cancer diagnosis is one method of disease management, which could also improve patient stratification and therapy response prediction, and thus reduce the mortality. Blood-based protein biomarkers, such as carcinoembryonic antigen (CEA) for colorectal carcinoma (CRC)2 and prostate specific antigen (PSA) for PCa3, have gained a lot of recognition with favorable disease outcome in the early diagnosis of these tumors. However, the use of these biomarkers in screening for early stages of PCa or CRC is still limited due to their low sensitivity and specificity as well as their inability to distinguish aggressive from indolent tumors, nor to distinguish benign or precancerous lesions from early-staged cancer 4, 5. Currently, there are no protein or other blood-based biomarkers are clinically available for early screening of BrCa or lung cancer (LuCa), and mammography or computed tomography (CT) are currently applied as the standard (or emerging) approach for BrCa and LuCa screening, respectively. Therefore, the development of novel and more sensitive biomarkers for early cancer diagnosis is needed to find supplement or complement approaches to the existing detection methods.
MicroRNAs (miRNAs) are small (~22 nucleotide), non-coding RNA molecules that have proven to be post-transcriptional regulators of gene expression by translational repression or degradation of targeted transcripts in a wide variety of biological processes including cell fate specification, proliferation, cell death, energy metabolism, and tumorigenesis6. The mature miRNA molecules circulating in plasma/serum are very stable due to the miRNA-Argonaute-protein complex7, 8, which provides the most important advantage of the non-invasive approach with remarkable stability and repeatability of measurement9. Since circulating miRNAs were detected in blood, they have been reported as an emerging class of blood-based biomarkers with the potential to provide information about distinct tumor biology in individual patients9, and aberrant levels of different circulating miRNAs species have been demonstrated as potential diagnostic or prognostic markers in various types of cancer, including lung10, 11, colorectal12, 13, prostate14, 15, and breast cancer 16, 17.
MicroRNA-21 (miR-21) is one of the first microRNA to be described as an oncomir, which targets on multiple tumor suppressors like PTEN18, PDCD419, p53 and TAp63 pathways20. Expression of miR-21 in tumor cells is up-regulated in a wide variety of solid tumors including prostate, colorectal, lung and breast cancer21, and elevated level of circulating miR-21 has been persistently observed in cancer patients from different populations by different researchers21-25. Furthermore, overexpression of serum or plasma miR-21 has been also found in other tumors including that of gastric26, biliary27, ovary28, cervix29, esophageal30, brain31, and liver32. These founding suggests that circulating miR-21 could be used as a useful blood-based diagnostic biomarker for various types of human cancers. However, the altered circulating miR-21 levels reported in these previous studies have mostly been observed by comparing miR-21 levels between cancer patients and healthy controls, with few comparing cancer patients with subjects of benign lesions. Thus, it still remains unclear whether circulating miR-21 can distinguish early-staged cancer patient form those subjects with benign lesions.
Recently, downregulated expression of microRNA-152 (miR-152) has been observed in various types of human cancer cell lines and tumor tissues including gastrointestinal cancers33, endometrial cancers34, ovarian cancer35, and hepatocellular carcinoma36, indicating that miR-152 might have the potential to act as a tumor suppressor in these tumors. Further studies have verified DNA methyltransferase 1 (DNMT1) as a target for miR-152 downregulation via a feedback mechanism of the CpG hypermethylation in the gene promoter in cancer cells 37-39. In prostate cancer, miR-152 can target TGFα to inhibit PCa cell migration and invasion and also interact with ERBB3 to contribute to PCa progression40, 41. A recent study has shown significantly differentiated level of circulating miR-152 in patients with non-small cell lung cancer (NSCLC), indicating it as a non-invasive biomarkers for prediction of recurrence in resectable (NSCLC) and the survival of squamous cell carcinoma patients42. An earlier study in urinary bladder cancer demonstrated that the RNA expression ratio of miRNA-126/miRNA-152 in urine sample enabled the detection of bladder cancer from urinary tract infections and healthy individuals43. However, no such study about the diagnostic value of plasma miRNA-152 has been reported in prostate, breast, colon or other human cancers so far.
In this study, to assess the diagnostic value of circulating miR-21 and miR-152 in cancer, we measured the level of plasma miR-21 and miR-152 in several cohorts of subjects including early staged cancer (I, II) patients for the four types of common cancers (breast, prostate, lung, and colon cancer), benign patients at the four organs, and healthy controls.
Patients and Methods
Patient Cohorts
Lung cancer: We enrolled a total of 143 plasma samples from 55 patients with non-small cell lung carcinoma (NSCLC), 35 patients with benign lesions and 53 random high risk controls at Rush University Medical Center (Chicago, IL) between 2004 and 2010. The early stage NSCLC patient inclusion criteria included the disease confined to the chest without evidence of distant metastases; no preoperative chemo- or radiotherapy within 1 year of our initial blood sampling; and a minimum of 2 years of clinical follow-up data. Patients with benign lesions include participants with a range of non-neoplastic pulmonary disorders (e.g. granulomas, hamartomas, and inflammatory lesions) as indicated in low-dose computed tomography (LDCT) screen. High risk subjects serving as normal controls in this study was defined to be individuals aged 55 to 75 years, having a smoking history of more than 30 pack-years, and having quit less than 15 years before randomization. All benign participants and high-risk individuals were followed with annual LDCT and remained cancer-free for a minimum 2-year follow-up. Cancer, benign and control samples were approximately age-, race-, gender-, and smoking status- matched as much as possible. Demographic information for these patients and controls is listed in Table 1. All patient data were acquired with written formal consent and in absolute compliance with the institutional review board (IRB) at Rush University Medical Center.
Breast cancer: Blood samples were obtained from The Cooperative Human Tissue Network (CHTN) Western Division and Southern Division, including 53 female sporadic breast cancer patients, 40 female patients with benign lesions, and 49 healthy females who served as normal controls. Cancer patient histopathology results were confirmed by surgical resection of the tumors and clinicohistopathological characteristics and tumor stage were assessed based on histobiopsy results. No preoperative chemotherapy or radiotherapy was applied to cancer patients included in this study. Breast benign lesions are defined as hyperplasia, fibroadenomas, cysts and some unspecified findings diagnosed at this organ. Control blood samples were collected from healthy women with no history of malignant diseases and no inflammatory conditions. All these cancer, benign and control samples were approximately age- and race- matched, as shown in Table 1. Rush University Medical Center IRB gave approval on the study with written consent for using all the subject information and biospecimens.
Prostate cancer: Unrelated men were recruited between the years 2001 and 2006 from the Division of Urology at Howard University Hospital in Washington, DC. Incident sporadic prostate cancer cases (n=65), with no family history, were identified by a urologist within the division or study coordinator and confirmed by a review of medical records. Clinical characteristics including Gleason grade, PSA, tumor-node metastasis stage, age at diagnosis, and family history were obtained for all cases from medical records. All these PCa patients were not given preoperative chemo- or radiotherapy. Disease aggressiveness was defined as ''low'' (T category <T2c and/or Gleason grade <8) or “high” (T category _T2c and/or Gleason grade _8). Benign subjects (n=51) were recruited from men diagnosed with benign prostatic hyperplasia (BPH) or high-grade prostatic intraepithelial neoplasm (HGPIN) lesions but without prostatic cancer. Healthy control subjects (n=74) unrelated to the cases and matched for age. Individuals who were ever diagnosed with benign prostate hyperplasia and/or had a prostate-specific antigen (PSA) value >2.5 ng/ml were not included as controls. All participants were between 40 and 85 years of age. The Howard University IRB approved the study and written consent was obtained from all participants.
Characteristics of cohorts of cancer patients, benign patients and normal controls. *Data are presented as median age at diagnosed.
| Lung | Colorectal | Breast | Prostate | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cancer (n=55) | Benign (n=35) | Normal (n=53) | Cancer (n=31) | Benign (n=33) | Normal (n=52) | Cancer (n=53) | Benign (n=40) | Normal (n= 49) | Cancer (n=65) | Benign (n=51) | Normal (n=74) | |
| Sex | ||||||||||||
| Male | 31 | 19 | 24 | 17 | 17 | 30 | 65 | 51 | 74 | |||
| Female | 24 | 16 | 29 | 14 | 16 | 22 | 53 | 40 | 49 | |||
| Race | ||||||||||||
| Caucasian | 53 | 34 | 50 | 20 | 24 | 34 | 50 | 40 | 42 | 35 | 26 | 43 |
| Noncaucasian | 2 | 1 | 3 | 11 | 9 | 18 | 3 | 7 | 30 | 25 | 31 | |
| Age*, yr | ||||||||||||
| Median | 67.42 | 59.74 | 60.06 | 63.71 | 59.62 | 59.06 | 53.04 | 47.27 | 60.78 | 62.01 | 65.22 | 62.14 |
| Range | 48-88 | 20-80 | 49-82 | 35-79 | 47-80 | 48-77 | 33-84 | 22-83 | 48-84 | 45-79 | 50-79 | 50-74 |
| Tumor stage | ||||||||||||
| Stage 1 | 43 | 26 | 19 | 37 | ||||||||
| Stage 2 | 12 | 5 | 34 | 28 | ||||||||
Colorectal cancer: Patients with CRC had a diagnosis determined at colonoscopy and confirmed by final surgical pathology at Rush University Medical Center. Early staged (Stage I or II) CRC patients without evidence of nodal disease were included (n=31). No any preoperative chemotherapeutical treatment was given to all CRC patient participants. Individuals (n=33) with adenomatous polyps were defined as benign patients according to CRC screening. Healthy volunteers (n=52) matched to age and race were individuals who underwent CRC screening by colonoscopy that was negative for either adenomatous polyps or CRC. The study was approved by the IRB of the Rush University Medical Center.
Plasma RNA extraction
The peripheral blood samples were collected using EDTA-anticoagulative tubes and centrifuged at 4000 RPM for 10 min, followed by a 15 min high-speed centrifugation at 12,000 RPM to completely remove cell debris. The supernatant serum was stored at -80°C until analysis. Total RNA was extracted from 200 μl of plasma using Qiagen miRNeasy Mini kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. In brief, the plasma was mixed with QIAzol Lysis Reagent and chloroform. After centrifugation at 12,000g at 4°C for 15 min, the aqueous phase was transferred into another tube, and 1.5 volumes of absolute ethanol were added. The mixture was then applied to miRNeasy Mini kit columns, following by washing with RWT and RPE buffers. The RNAs were finally eluted in 40 μl of RNase-free water. For normalization of sample-to-sample variation during RNA extraction and as internal control, same amounts of 25 fmol a synthetic C. elegans miRNA-39 (Cel-miR-39) was added into each plasma mixture.
Quantitative PCR
MiRNAs were measured using Taqman miRNA assay kits (Applied Biosystems, USA) according to the manufacturer's protocol. Briefly, about 30 ng enriched RNA was reverse transcribed with a TaqMan microRNA Reverse Transcription Kit (Applied Biosystems, USA) in a 15 µL reaction volume. Expression levels of miR-21 and miR-152 were quantified in triplicate by qRT-PCR using human TaqMan MicroRNA Assay Kits (Applied Biosystems, USA) on Eppendorf iplex 4 system (Eppendorf North America, Hauppauge, NY). Spiked-in Cel-miR-39 was used as a normalizer for plasma miRNA quantification.
Statistical analysis
The relative expression of miR-21 and miR-152 was analyzed by using the 2- Δ Δ Ct method, as previously described9. The Mann-Whitney U test was used to compare the expression of plasma miRNAs between the different groups. The Younden index determined the threshold for the plasma miRNA concentrations. The correlation between clinicopathologic features and plasma miRNA levels was determined by Student's t-test or Χ2 test. All tests were 2-sided and a P value less than 0.05 (95% CI) was considered statistically significant. Statistical analysis was performed using SPSS 16.0 software (SPSS Ltd., UK).
Results
Patient Population
In this study a total of 591 participants were recruited, including 204 cancer patients (55 LuCa, 31 CRC, 53 BrCa and 65 PCa), 159 benign lesions (35 lung benign nodules, 33 advanced colon adenomas, 40 breast benign lesions and 51 BPHs/HGPINs), and 228 normal subjects (Table 1). For each type of cancer, only early-staged patients (stage I or II) were selected. The independent normal subjects were recruited separately and matched in race, sex and age for each type of cancer and benign lesions. The cohort of normal subjects for lung cancer was also described as a “high-risk” population, in which all the healthy subjects have had a smoking history of more than 30 pack-years and quit less than 15 years. Colorectal advanced adenomas, which is considered as precancerous lesion, was also included in the group of “benign lesions” in this study. Only matched healthy females and males were included in the cohort of normal subjects for BrCa and PCa, without significant difference in age or race when compared to the patient groups, respectively.
Evaluation of plasma miR-21 and miR-152 expression level in cancer patients
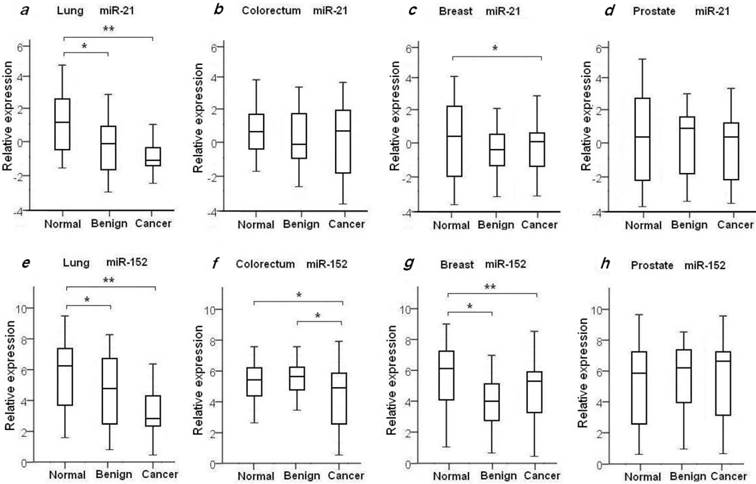
To evaluate the diagnostic value of oncomir miR-21 for the four common types of cancer, the levels of plasma miR-21 were measured in all 204 recruited cancer patients with LuCa, CRC, BrCa and PCa, and in the four independent normal control cohorts, as shown in Table 2 and Fig. 1. We found the mean relative level of plasma miR-21 was increased in LuCa patients with 2.39-fold change when compared to high-risk controls (p=1.07×10-4) (Fig. 1a), and 1.92-fold change in BrCa patients with comparison to the matched healthy females (p=0.03) (Fig. 1c. We also observed a slight up-regulation of miR-21 in CRC (1.23-fold, Fig. 1b) and PCa patients (1.21-fold, Fig. 1d), with no significant differences when compared to healthy controls (Table 2).
It has been previously shown that miR-152 functions as a tumor suppressor with evidence of decreased expression in various types of tumors37-39. To investigate whether plasma miR-152 is differentiated in the selected four common types of cancers, we measured the expression level of circulating miR-152 among cancer patients, benign individuals and healthy controls. Unexpectedly, we observed increased level of miR-152 in cancer patients of LuCa, CRC and BrCa when compared to normal controls (Figs. 1e-h). As shown in Table 2 and Fig. 1, significant up-regulated miR-152 expression was observed with 2.68-fold change in LuCa patients (p=1.52×10-4) (Fig. 1e), 2.03-fold in CRC (p=0.02) (Fig. 1f), and 2.91-fold in BrCa (p=2.75×10-3) (Fig. 1g), when compared to normal controls, respectively.
Up-regulation of plasma miR-21 miR-152 expression in patients with benign lesions
We also examined the plasma miR21 and miR-152 expression levels in the selected four groups of benign patients. We did not observe significant difference of plasma miR-21 expression in benign patients as compared to cancer patients in all four selected types of diseases (Table 2, Figs. 1a-d). As compared with normal controls, significant higher level of miR-21 was observed in lung benign patients (p=4.42×10-3), while no difference was observed between benign patients and normal individuals in the other three types of disease (Table 2, Figs. 1a-d).
Compared to normal controls, elevated levels of miR-152 were also observed in lung benign patients with 1.98-fold change (p=0.02) (Fig. 1e), and in breast benign patients with 3.41-fold change (p=2.3×10-4) (Fig. 1g), respectively. No significant difference of miR-152 expression was seen between prostate and colorectum benign patients and their matched normal subjects (Figs. 1f and 1h). Interestingly, we noticed significant difference of miR-152 in CRC patients and patients with adenomatous polyps (Table 2, Fig. 1f).
Expression changes of plasma miR-21 and miR-152 in early stage cancer cases, benign patients and healthy controls. Bold indicates statistical significance of expression level with * for p<0.05 and ** for p<0.001.
| miRNAs | Cancer vs. Normal | Cancer vs. Benign | Benign vs. Normal | |||
|---|---|---|---|---|---|---|
| Fold change | P value | Fold change | P value | Fold change | P value | |
| miR-21 | ||||||
| Lung | 2.39 | 1.07E-04** | 1.14 | 0.29 | 2.10 | 4.42E-03 |
| Colorectal | 1.23 | 0.28 | 0.99 | 0.49 | 0.96 | 0.22 |
| Breast | 1.92 | 0.03* | 0.88 | 0.31 | 1.69 | 0.07 |
| Prostate | 1.21 | 0.37 | 1.41 | 0.23 | 0.85 | 0.38 |
| miR-152 | ||||||
| Lung | 2.68 | 1.52E-04** | 1.38 | 0.15 | 1.98 | 0.02* |
| Colorectal | 2.03 | 0.02* | 1.99 | 0.01* | 1.02 | 0.46 |
| Breast | 2.91 | 2.75E-03* | 0.76 | 0.19 | 3.41 | 2.33E-04** |
| Prostate | 1.16 | 0.41 | 1.26 | 0.33 | 0.91 | 0.28 |
Plasma expression levels of miR-21 and miR-152 in 4 types of cancer patients, benign patients and normal controls. The relative expression level of miR-21 and miR-152 was determined as ΔCt = CtmiR-21 or CtmiR-152 - CtCel-miR-39. Statistical significance of miRNAs expression level was asterisked as * for p<0.05 and ** for p<0.001.
The diagnostic value of miR-21 and miR-152 for cancer early detection
To further evaluate the diagnostic efficiency of plasma miR-21 and miR-152 in discriminating patients with early-staged cancer from healthy individuals and benign patients, receiver-operator characteristic (ROC) analyses were applied in these different groups between which statistically significant expression level of plasma miR-21 or miR-152 was observed. As shown in Fig. 2, the differentiated miR-21 was found to be able to distinguish cancer patients with LuCa or BrCa from matched normal controls, with area under the receiver-operator characteristic (AUC) values of 0.701 for lung cancer (Fig. 2a) and 0.613 for breast cancer (Fig. 2c), respectively. Comparatively, miR-152 exhibited AUC values of 0.701 for lung cancer (Fig. 2b) and 0.687 for breast cancer (Fig. 2d) in the discrimination of cancer from normal individuals. Furthermore, miR-152 was also able to distinguish CRC patients from the matched healthy individuals with AUC of 0.539 (Fig. 2e). Neither miR-21 nor miR-152 was observed to be able to discriminate prostate cancer patients from the matched healthy controls in this presented study.
ROC analyses for distinguishing cancer from normal controls and benign lesions in LuCa, BrCa and CRC using miR-21 and miR-152. (a) miR-21 for LuCa vs. normal, (b) miR-152 for LuCa vs. normal, (c) miR-21 for BrCa vs. normal, (d) miR-152 for BrCa vs. normal, (e) miR-152 for CRC vs. normal, and (f) miR-152 for CRC vs. polyps.
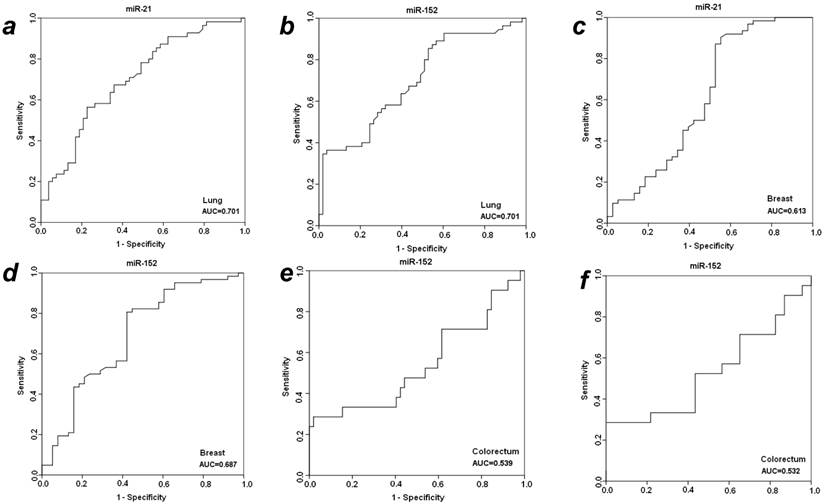
With significantly differentiated level in cancer CRC patients as compared to patients with adenomatous polyps, plasma miR-152 was considered to be able to distinguish CRC from benign colorectal polyp lesions, with a slight AUC of 0.537 (Fig. 2f) exhibited in the cohort of 31 cancer patients. No such observation was obtained for miR-152 in the other three types of cancers when compared to benign patients. The expression level of plasma miR-21 was not significantly changed when compared between cancer vs. benign groups in all the four types of cancers.
Discriminating efficiency of plasma miR-21 and miR-152 between cancer and non-cancer
To further evaluate the diagnostic value of plasma miR-21 and miR-152 in distinguishing cancer from benign and normal individuals, we analyzed the expression level of these two miRNAs by re-grouping these subjects. We first grouped benign patients together with matched normal individuals as a “non-cancer” group, and then compared the expression level of plasma miR-21 and miR-152 between cancer vs. non-cancer group for all the four types of cancers (Table 3). Significant higher level of plasma miR-21 was observed with 1.85-fold change in lung cancer patients as compared to non-cancer groups (p<0.001). The relative AUC for miR-21 in discrimination of lung cancer from non-cancer was 0.645 (Fig. 3a). However, no such significant alteration was found for miR-21 between cancer and non-cancer groups in the other three types of cancers (Table 3). On the other hand, the elevated expression level of miR-152 was observed significantly in cancer group when compared to non-cancer individuals with about 2-fold change (p<0.01) for both LuCa and CRC (Table 3), and the corresponding AUC values were 0.652 for LuCa and 0.537 for CRC, respectively (Figs. 3b and 3c).
Comparison of miR-21 and miR-152 expression between different status cancers vs non-cancers and diseases vs non-diseases. Bold indicates statistical significance of expression level with * for p<0.01 and ** for p<0.001.
| Types | Cancer vs. Non-cancer | Disease vs. Normal | ||
|---|---|---|---|---|
| Fold change | P value | Fold change | P value | |
| miR-21 | ||||
| Lung | 1.85 | 2.44E-03* | 2.28 | 4.37E-05** |
| Colorectal | 1.16 | 0.32 | 1.23 | 0.23 |
| Breast | 1.04 | 0.07 | 1.82 | 0.02* |
| Prostate | 1.3 | 0.29 | 0.99 | 0.44 |
| miR-152 | ||||
| Lung | 2.07 | 1.52E-03* | 2.39 | 1.84E-04** |
| Colorectal | 2.01 | 2.67E-03* | 1.41 | 0.08 |
| Breast | 1.38 | 0.12 | 2.93 | 1.63E-04** |
| Prostate | 1.14 | 0.35 | 0.96 | 0.48 |
ROC analyses for miR-21 and miR-152 in discriminating cancer vs. non-cancer and disease vs. normal. (a) miR-21and (b) miR-152 for LuCa vs. Non-cancer, (c) miR-152 for CRC vs normal, (d) miR-21 and (e) mir-152 for “lung disease” vs. normal, (f) miR-21 and (g) mir-152 for “breast disease” vs. normal. Non-cancer, combined group of normal controls and benign patients. Disease, combined group of cancer patients and benign patients.
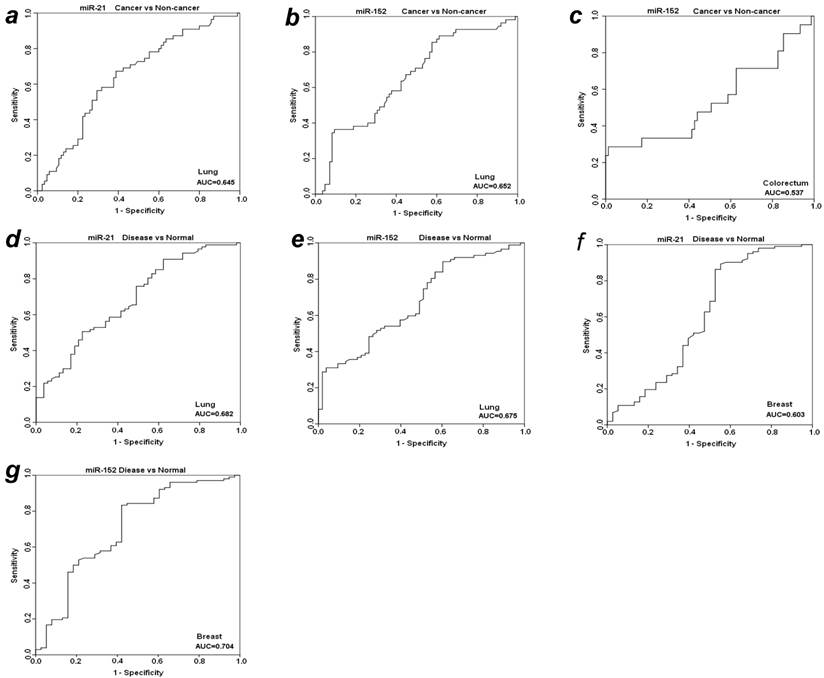
Since there was no significant difference of plasma miR21 and miR-152 between cancer groups and benign patient groups in almost the four types of cancers (except miR-152 in cancer vs benign in CRC) (Table 2), we combined cancer patients and benign patients into a “disease” group, and compared the expression level of these two miRNAs between “disease” and normal controls. Significant differences were observed for both miR21 and miR-152 in lung and breast “disease” group (all p<0.05), but not in colorectal or prostate disease (Table 3). The AUCs of miR-21 and miR-152 in distinguishing disease from normal were 0.682 and 0.674 for lung (Figs. 3d and 3e), 0.603 and 0.704 for breast (Figs. 3f and 3g), respectively.
Discussion
In this present study, we evaluated plasma miR-21 and mir-152 as potential diagnostic biomarkers for early detection of multiple types of cancer. Our results confirmed significantly elevated miR-21 level in lung cancer patients and breast patients, but not in CRC or PCa patients. Unexpectedly, we observed up-regulated expression of plasma miR-152 in lung, breast, and colorectal cancer. To our knowledge, this is the first report to reveal the association of plasma miR-152 level with multiple types of cancer. We also investigated the expression level of miR-21 and miR-152 in the four groups of patients with benign or precancerous lesions at one of lung, colorectum, breast or prostate sites. Our results indicated significant lower level of miR-152 in patients with adenomatous polyps as compared to CRC patients, but not in the other three benign patients groups. In addition, no significant difference of mir-21 expression was observed between benign patients and cancer patients in all the four selected types of cancer.
Currently, published studies on circulating microRNAs as cancer detection biomarkers, mostly focusing on a specific type of cancer, have identified a wide variety of microRNAs for multiple types of cancers. However, results from these studies may lead to misunderstanding of the identified miRNAs as potential biomarkers specific for a certain type of cancer. For example, miR-21 was such a miRNA candidate identified in many independent studies, in each of them it was suggested as a biomarker for a certain disease by the researcher. Therefore, simultaneous assessment of a cancer-related miRNA marker in various types of cancer and disease status could improve the sensitivity and specificity of this marker for cancer prediction. In this study, we evaluated miR-21 in the four most common types of cancer and the corresponding benign lesions, and our results suggest that miR-21 was a non-specific biomarker for early cancer screening, which was not able to distinguish malignancy from non-cancerous benign lesions.
The oncomir miR-21 is believed to play a role in a wide variety of cancers, and high level of plasma or serum miR-21 has been reported in various cancers21-25. It is well known that miR-21 is a secreted miRNA which is derived from exosomes44, 45. When tumors actively release exosomes into peripheral circulation, the miRNA molecule contained in exosomes would act as a diagnostic biomarker for cancer44, 45. Interestingly, it is shown that postoperative reductions in circulating miR-21 levels occurred exclusively among cancer patients with potentially curative surgeries or chemotherapy treatment, indicating that circulating miR-21 would also serve as a long-term follow-up biomarker in cancer prognosis46-48. Moreover, miR-21 has also been shown to be involved in development of other human diseases, including heart disease with observation of significantly down-regulated miR-21 expression in acute myocardial infarcted areas49, 50. We also observed higher expression level of plasma miR-21 in benign diseases in lung and breast in the cohorts of samples we used in this study. These observations altogether indicated that miR-21 may be involved in multiple types of diseases, inflammatory process, and responses to chemotherapeutical drugs, and therefore the circulating miR-21 may not be an ideal specific biomarker for the detection of cancer.
MiR-152 is a member of the miR-148/152 family33. Like the other two members, miR-148a and miR-148b, miR-152 is involved in the growth and development of normal tissues, as well as in the genesis and development of disease51. It has been shown that miR-152 functions as a tumor suppressor, and its expression is decreased in various tumor tissues 33-39. Surprisingly, increased level of plasma miR-152 was observed in lung, breast and colon cancer patients and in patients with lung or breast benign lesions, as compared to matched normal controls in this study (Table 2 and Fig. 1). Unlike the oncomir miR-21, which is high expressed in both tumor tissues and in blood of cancer patients, this observation of increased miR-152 level in plasma samples from various cancer patients was unexpected because miR-152 is believed to be down-regulated in the tumor tissues. Actually, recent studies have supported the “inconsistency” of high circulating miR-152 level in cancer patients with several pieces of evidence in bladder cancer and lung cancer 42, 52. In lung cancer, plasma miR-152 expression level was indicated to significantly predict survival of squamous cell carcinoma, with a low plasma miR-152 level associated with poor disease-free survival of NSCLC patients42. However, a very recent study in bladder cancer revealed that high circulating miR-152 level was presented in bladder cancer patients which was independently associated with tumor recurrence of nonmuscle-invasive bladder cancer and worse recurrence-free survival52. As this is the first report on high circulating miR-152 level in early-staged cancer of lung, breast, colorectal, and prostate, further studies are needed to confirm in our findings in larger size of samples. Moreover, it will be worthy to further investigate the underlying mechanism of this non-coincidence of miR-152 high level in plasma and its low presence level in tumor tissues.
In this present study, we observed slight to medium predictive efficiency of miR-21 and miR-152 in discrimination of LuCa, CRC, and BrCa from normal controls, with little valuable power in distinguishing cancer from benign lesions. We realized that the size of subjects including cancer patients, benign patients and normal controls were still small for each type of cancer, limiting the evaluation on miR-21 and miR-152 as predictive biomarker in early detection of cancer. However, we still believed that the nature of miR-21 and miR-152 expression in plasma which was revealed in the multiple cohorts of samples is the key point for that. Therefore, searching for more specific and powerful miRNA biomarkers to discriminate early cancer from precancerous lesions for these common types of cancer using novel strategies is still warranted.
Conclusions
In this study, we detected the plasma miR-21 and miR-152 expression level in patients with lung, breast, colorectal and prostate cancer, with comparison to that in matched normal controls and patients with benign lesions at the respective organ sites. Our results showed the increasing levels of plasma miR-152 and miR-21 in some types of cancer patients and in some patients with benign lesions, with limited predictive value in discriminating cancer from healthy controls and benign lesions. Further studies aiming at searching for more specific and sensitive miRNA biomarkers for early detection of each types of cancer, especially in distinguishing cancer from benign lesions, are still needed.
Acknowledgements
We give thanks to all collaborators who were involved in the sample collection and preparation. This project is supported by the NIH grant (1R21 CA164764) and Bears Care Foundation to Youping Deng.
Competing Interests
The authors declare no conflict of interest.
References
1. Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9-29
2. Jain P, Mondal SK, Sinha SK, Mukhopadhyay M, Chakraborty I. Diagnostic and prognostic significance of different mucin expression, preoperative CEA, and CA-125 in colorectal carcinoma: A clinicopathological study. J Nat Sci Biol Med. 2014;5:404-8
3. Obort AS AM, Akinloye O. Prostate-specific antigen: any successor in sight? Rev Urol. 2013;15:97-107
4. Gomella LG, Liu XS, Trabulsi EJ, Kelly WK, Myers R, Showalter T, Dicker A, Wender R. Screening for prostate cancer: the current evidence and guidelines controversy. Can J Urol. 2011;18:5875-83
5. Hanash SM, Baik CS, Kallioniemi O. Emerging molecular biomarkers-blood-based strategies to detect and monitor cancer. Nat Rev Clin Oncol. 2011;8:142-50
6. Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9:831-42
7. Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223-33
8. Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003-8
9. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N. et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513-8
10. Zheng D, Haddadin S, Wang Y, Gu LQ, Perry MC, Freter CE, Wang MX. Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int J Clin Exp Pathol. 2011;4:575-86
11. Cazzoli R, Buttitta F, Di Nicola M, Malatesta S, Marchetti A, Rom WN, Pass HI. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol. 2013;8:1156-62
12. Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118-26
13. Redova M, Sana J, Slaby O. Circulating miRNAs as new blood-based biomarkers for solid cancers. Future Oncol. 2013;9:387-402
14. Srivastava A, Suy S, Collins SP, Kumar D. Circulating MicroRNA as Biomarkers: An Update in Prostate Cancer. Mol Cell Pharmacol. 2011;3:115-24
15. Bryant RJ, Pawlowski T, Catto JW, Marsden G, Vessella RL, Rhees B, Kuslich C, Visakorpi T, Hamdy FC. Changes in circulating microRNA levels associated with prostate cancer. Br J Cancer. 2012;106:768-74
16. Ng EK, Li R, Shin VY, Jin HC, Leung CP, Ma ES, Pang R, Chua D, Chu KM, Law WL, Law SY, Poon RT. et al. Circulating microRNAs as specific biomarkers for breast cancer detection. PLoS One. 2013;8:e53141
17. Cuk K, Zucknick M, Heil J, Madhavan D, Schott S, Turchinovich A, Arlt D, Rath M, Sohn C, Benner A, Junkermann H, Schneeweiss A. et al. Circulating microRNAs in plasma as early detection markers for breast cancer. Int J Cancer. 2013;132:1602-12
18. Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647-58
19. Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128-36
20. Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164-72
21. Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP. et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065-70
22. Wang F, Zheng Z, Guo J, Ding X. Correlation and quantitation of microRNA aberrant expression in tissues and sera from patients with breast tumor. Gynecol Oncol. 2010;119:586-93
23. Zhang HL, Yang LF, Zhu Y, Yao XD, Zhang SL, Dai B, Zhu YP, Shen YJ, Shi GH, Ye DW. Serum miRNA-21: elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate. 2011;71:326-31
24. Shen J, Todd NW, Zhang H, Yu L, Lingxiao X, Mei Y, Guarnera M, Liao J, Chou A, Lu CL, Jiang Z, Fang H. et al. Plasma microRNAs as potential biomarkers for non-small-cell lung cancer. Lab Invest. 2011;91:579-87
25. Kanaan Z, Rai SN, Eichenberger MR, Roberts H, Keskey B, Pan J, Galandiuk S. Plasma miR-21: a potential diagnostic marker of colorectal cancer. Ann Surg. 2012;256:544-51
26. Cui L, Zhang X, Ye G, Zheng T, Song H, Deng H, Xiao B, Xia T, Yu X, Le Y, Guo J. Gastric juice MicroRNAs as potential biomarkers for the screening of gastric cancer. Cancer. 2013;119:1618-26
27. Kishimoto T, Eguchi H, Nagano H, Kobayashi S, Akita H, Hama N, Wada H, Kawamoto K, Tomokuni A, Tomimaru Y, Umeshita K, Doki Y. et al. Plasma miR-21 is a novel diagnostic biomarker for biliary tract cancer. Cancer Sci. 2013;104:1626-31
28. Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, Calin GA, Menard S. et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699-707
29. Lui WO, Pourmand N, Patterson BK, Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67:6031-43
30. Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, Morimura R, Nagata H, Kosuga T, Iitaka D, Konishi H, Shiozaki A, Fujiwara H, Okamoto K. et al. Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer. 2011;105:104-11
31. Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029-33
32. Tomimaru Y, Eguchi H, Nagano H, Wada H, Kobayashi S, Marubashi S, Tanemura M, Tomokuni A, Takemasa I, Umeshita K, Kanto T, Doki Y. et al. Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. J Hepatol. 2012;56:167-75
33. Chen Y, Song Y, Wang Z, Yue Z, Xu H, Xing C, Liu Z. Altered expression of MiR-148a and MiR-152 in gastrointestinal cancers and its clinical significance. J Gastrointest Surg. 2010;14:1170-9
34. Tsuruta T, Kozaki K, Uesugi A, Furuta M, Hirasawa A, Imoto I, Susumu N, Aoki D, Inazawa J. miR-152 is a tumor suppressor microRNA that is silenced by DNA hypermethylation in endometrial cancer. Cancer Res. 2011;71:6450-62
35. Zhou X, Zhao F, Wang ZN, Song YX, Chang H, Chiang Y, Xu HM. Altered expression of miR-152 and miR-148a in ovarian cancer is related to cell proliferation. Oncol Rep. 2012;27:447-54
36. Huang J, Wang Y, Guo Y, Sun S. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology. 2010;52:60-70
37. Ji W, Yang L, Yuan J, Zhang M, Qi D, Duan X, Xuan A, Zhang W, Lu J, Zhuang Z, Zeng G. MicroRNA-152 targets DNA methyltransferase 1 in NiS-transformed cells via a feedback mechanism. Carcinogenesis. 2013;34:446-53
38. Xiang Y, Ma N, Wang D, Zhang Y, Zhou J, Wu G, Zhao R, Huang H, Wang X, Qiao Y, Li F, Han D. et al. MiR-152 and miR-185 co-contribute to ovarian cancer cells cisplatin sensitivity by targeting DNMT1 directly: a novel epigenetic therapy independent of decitabine. Oncogene. 2014;33:378-86
39. Theodore SC, Davis M, Zhao F, Wang H, Chen D, Rhim J, Dean-Colomb W, Turner T, Ji W, Zeng G, Grizzle W, Yates C. MicroRNA profiling of novel African American and Caucasian Prostate Cancer cell lines reveals a reciprocal regulatory relationship of miR-152 and DNA methyltranferase 1. Oncotarget. 2014;5:3512-25
40. Zhu C, Li J, Ding Q, Cheng G, Zhou H, Tao L, Cai H, Li P, Cao Q, Ju X, Meng X, Qin C. et al. miR-152 controls migration and invasive potential by targeting TGFalpha in prostate cancer cell lines. Prostate. 2013;73:1082-9
41. Lichner Z, Fendler A, Saleh C, Nasser AN, Boles D, Al-Haddad S, Kupchak P, Dharsee M, Nuin PS, Evans KR, Jung K, Stephan C. et al. MicroRNA signature helps distinguish early from late biochemical failure in prostate cancer. Clin Chem. 2013;59:1595-603
42. Sanfiorenzo C, Ilie MI, Belaid A, Barlesi F, Mouroux J, Marquette CH, Brest P, Hofman P. Two panels of plasma microRNAs as non-invasive biomarkers for prediction of recurrence in resectable NSCLC. PLoS One. 2013;8:e54596
43. Hanke M, Hoefig K, Merz H, Feller AC, Kausch I, Jocham D, Warnecke JM, Sczakiel G. A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol Oncol. 2010;28:655-61
44. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654-9
45. Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13-21
46. Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105:849-59
47. Yang Y, Qian J, Chen Y, Pan Y. Prognostic role of circulating microRNA-21 in cancers: evidence from a meta-analysis. Tumour Biol. 2014;35:6365-71
48. Chen J, Wang X. MicroRNA-21 in breast cancer: diagnostic and prognostic potential. Clin Transl Oncol. 2013;16:225-33
49. Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J. et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980-4
50. Dong S, Cheng Y, Yang J, Li J, Liu X, Wang X, Wang D, Krall TJ, Delphin ES, Zhang C. MicroRNA expression signature and the role of microRNA-21 in the early phase of acute myocardial infarction. J Biol Chem. 2009;284:29514-25
51. Chen Y, Song YX, Wang ZN. The microRNA-148/152 family: multi-faceted players. Mol Cancer. 2013;12:43
52. Jiang X, Du L, Wang L, Li J, Liu Y, Zheng G, Qu A, Zhang X, Pan H, Yang Y, Wang C. Serum microRNA expression signatures identified from genome-wide microRNA profiling serve as novel noninvasive biomarkers for diagnosis and recurrence of bladder cancer. Int J Cancer. 2014
Author contact
![]() Corresponding author: Youping Deng, Ph.D., Department of Internal Medicine and Biochemistry, Rush University Medical Center. 630 S. Hermitage Ave. Kidston House, Room 408. Chicago, IL 60612. Tel: 312-563-4440, Fax: 312-563-4484. Email: Youping_dengedu.
Corresponding author: Youping Deng, Ph.D., Department of Internal Medicine and Biochemistry, Rush University Medical Center. 630 S. Hermitage Ave. Kidston House, Room 408. Chicago, IL 60612. Tel: 312-563-4440, Fax: 312-563-4484. Email: Youping_dengedu.

 Global reach, higher impact
Global reach, higher impact