3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2015; 6(4):302-309. doi:10.7150/jca.11036 This issue Cite
Research Paper
Association between CYP1A1 Ile462Val Polymorphism and Oral Squamous Cell Carcinoma Susceptibility: Evidence from 13 Investigations
1. Department of General Medicine, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou 325000, Zhejiang, China
2. Department of Oral and Maxillofacial Surgery, Peking University School and hospital of Stomatology, Beijing 100081, China
* Xiao-Lei Yang and Shang Xie contributed equally to this work.
Received 2014-11-11; Accepted 2014-12-16; Published 2015-1-22
Abstract
CYP1A1 Ile462Val polymorphism might play a key role in pathogenesis of oral squamous cell carcinoma (OSCC). Many case-control studies have investigated the association between CYP1A1 Ile462Val polymorphism and OSCC susceptibility. However, the conclusions are inconsistent. To aim a convincible conclusion, we carried out a meta-analysis to systematically evaluate the association of CYP1A1 Ile462Val polymorphism with OSCC susceptibility. We searched Pubmed, Web of Science, Ovid and Embase databases for available publications. The odds ratio (OR) with the corresponding 95% confidence interval (95% CI) was carried out to estimate the association. A total of 13 papers including 1468 cases and 2183 controls were included, a significant increased OSCC risk was observed in recessive model (OR=1.64, 95% CI=1.08-2.49), but not other genetic models. Our results suggest that the homozygous variant of CYP1A1 Ile462Val might be a risk factor of OSCC.
Keywords: CYP1A1, polymorphism, OSCC, meta-analysis
Introduction
Oral cancer is one of the most commonly occurred cancers worldwide and causes a considerable problem to human health being a high mortality rates and disfigurement [1, 2]. About 90% of oral malignant neoplasms are oral squamous cell carcinomas (OSCC), followed by adenocarcinoma and, rarely, other types [3]. Despite advances in treatment for OSCC, the 5-year survival rate remains undesirable [4-6]. Hence, investigating the risk factors and developing the early diagnosis for treatment and prevention of OSCC are urgently needed.
Epidemiological studies showed that OSCC have been associated with high tobacco abuse and alcohol consumption [7-11]. However, not all individuals who have smoking and alcohol habits develop this fatal disease, suggesting that individual genetic factor may also be involved in disease etiology. The research results of human genome project (HGP) have demonstrated that different individuals are all 99.9% the same genes and the tiny genetic difference sequences are mainly gene single nucleotide polymorphisms (SNPs). Therefore, inter-individual differences in expression of SNPs might contribute to variability in risk towards various types of cancers including OSCC. Currently, the published evidences showed that there were significant associations of gene polymorphisms with the susceptibility of many cancers, such as Glutathione S-transferase (GSTs) and Cytochrome P4501 A1 (CYP1A1) gene polymorphisms with lung of squamous cell carcinoma, polymorphism of 8q24 rsl3281615 with risk of breast cancer, CYP1A1 and GSTs gene polymorphisms with head and neck cancer [12-19]. However, the associations of CYP1A1 Ile462Val variant with OSCC risks are inconsistent. For example, some studies [20-22] reported that CYP1A1*2C polymorphism was a risk factor of OSCC, however, some studies [23, 24] opposed it.
CYP1A1 is a member of the CYP family that participates in the metabolism of xenobiotics and endogenous compounds, encoding for the aryl hydrocarbon hydrolase (AHH), which is involved in activation of polycyclic aromatic hydrocarbon (PAHs) and aromatic amines and is expressed in oral tissue [25]. Because CYP1A1 is able to activate carcinogenic PAHs and its expression and function are affected by gene polymorphisms, people pay more attention to the association of cancer and CYP1A1. According to the published studies, the CYP1A1 have several single nucleotide polymorphisms, which may alter the activities of their enzymes and increase carcinogen activation and yield to carcinogenicity. The CYP1A1*2C or CYP1A1 exon7 or Ile462Val is one of the most common polymorphisms, which is a transition from valine to isoleucine at codon 462 in the exon7 region resulting in the introduction of an BsrDI restriction site and associating with increase in enzyme activity and hence affecting the risks of carcinoma [26, 27]. Exon7 restriction site polymorphism brings about three genotypes, wild type (Val/Val), heterozygous variant (Val/Ile) and homozygous variant (Ile/Ile) [28].
Considering the significance of CYP1A1 Ile462Val polymorphism in the occurrence and development of malignancies including OSCC, we systematically evaluated the association between CYP1A1 Ile462Val polymorphism and OSCC susceptibility through a meta-analysis.
Materials and Methods
Search strategy
We searched the following databases: Pubmed, Web of Science, Ovid and Embase databases without language limitations, and the last research was updated on August 8, 2014. The search process was designed to find primarily all relevant articles and the search strategies were listed as follows: (1) Cytochrome P450 1A1 or P450 1A1 or CYP1A1 or CYP1A1∗2C or exon7 or Ile462Val; and (2) genotype or polymorphism or allele or variant; and (3) oral squamous cell carcinoma or OSCC or mouth neoplasm or oral cancer or oral carcinoma or oral tumor. Searched results were in dependently screened by two authors according to the titles, abstracts and types of articles, and irrelevant papers were dropped out. Manual review of the references cited in the selected articles was undertaken to obtain articles that might have been missed in the search process. Then we downloaded the relevant papers and further screened to identify potentially eligible studies. If essential data were not provided in the original articles, every effort was made to contact the authors.
Inclusion/exclusion criteria
All relevant case-control studies were screened, irrespective of languages. In the meta-analysis, the following criteria were set and reviewed by two independent authors (Xiao-Lei Yang and Shang Xie): (1) studies should be concerned about the relation of CYP1A1 Ile462Val polymorphism with OSCC risk, and cases of identification of OSCC were histologically confirmed; (2) each trial should be an observational study (case-control or cohort), and the subjects should be human beings; (3) papers must offer the size of the sample, and the genetic distribution or the original information that can help infer the relevant results; (4) when multiple publications from a particular research group reported data from overlapping samples, the study reporting the largest or latest dataset was included.
Exclusion criteria: (1) review articles or editorials or meta-analysis; (2) case reports, or lack of case-control studies; (3) studies that estimated the risk of secondary tumors, prognosis, recurrence, or response to treatment. For conflicting evaluation, an agreement was reached following a discussion. If a consensus could not be attained, another author was invited to solve the dispute and then a final result was made by the majority of the votes. After rigorous searching, we viewed all papers in accordance with the criteria defined above for further analysis.
Data extraction
All data was independently reviewed and extracted from the included papers by two investigators. Differences between reviewers were solved by discussion, or through consultation if necessary. The following characteristic were collected from each study, ethnicity, country, sample size, control source, matching contents, the Hardy-Weinberg Equilibrium (HWE), and genetic distribution of cases and controls. If there are studies deviating from HWE, we will increase the analysis of subgroup based on the HWE. When the data were not clear nor presented by the author in the publication, contacting them for further details were attempted.
Quality assessment
We performed the Newcastle-Ottawa Scale (NOS) [29] to estimate the methodological quality of included studies. The NOS system categorizes into three dimensions including selection, comparability, and ascertainment of outcome. A star system is used to assess the quality of all selected studies. The scores of NOS ranges from zero (the lowest) to nine (the highest) stars, with more stars indicating a better methodological quality. The assessment was performed independently by two investigators and the inconformity was solved by a discussion, or consultation if necessary.
Statistical analysis
All the data management and analysis for this meta-analysis were performed with STATA 11.0 software (Stata corporation, College Station, TX). The odds ratios (ORs) with corresponding 95% confidence intervals (CIs) were used to estimate the association between CYP1A1 Ile462Val polymorphism and OSCC risk. In order to calculate heterogeneity of studies, the Chi-Square test was used and significance was set at P value less than 0.05 level [30]. The inconsistency index I2 was calculated to assess the variation caused by heterogeneity. If the P value of the heterogeneity test was >0.10 and I2<25%, the fixed-effects model was performed to calculate the combined OR, which assumed the same homogeneity of effect size across all studies. If the P value of the heterogeneity test was <0.10 and I2>25%, it showed that the between-study heterogeneity was statistically significant, and a random effect model was used to estimate the pooled OR. The funnel plot was used to test the underlying publication bias, and the funnel plot asymmetry was estimated by Egger's linear regression [31]. Sensitivity analyses were performed to identify the influence of the individual studies on the combined OR. In the analysis, we excluded each study to assess if stability between the remaining studies was reached.
Trial sequential analysis (TSA)
According to the Cochrane Handbook, meta-analyses and systematic reviews are considered to be the best available evidence if all eligible trials are included. However, 'the best available evidence' might not be always equal to 'sufficient evidence'. Based on this issue, we applied the TSA to estimate the power of the current conclusions [32-34]. The methods of using TSA were based on the 'User manual for Trial Sequential Analysis (TSA)'.
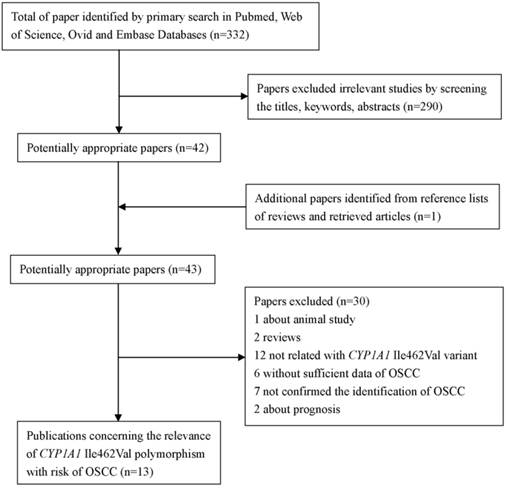
The flow diagram of searching process
Results
Characteristics of included studies
As shown in Figure 1, a total of 332 studies were retrieved by the literature search. Of which, 290 studies were excluded as being irrelevant to CYP1A1 Ile462Val or OSCC or genetic polymorphism or not human studies. And one potential eligible article was obtained by screening the references of reviews. After more detailed assessment for the left 43 potential eligible articles, one article was excluded for animal study; two papers were reviews. Following this, 12 papers were concerned about CYP1A1*2A or other genes, but not CYP1A1 Ile462Val. Besides, six papers were excluded because of failing to provide sufficient data. In addition, seven studies were excluded because the cases just were diagnosed oral cancer, were not confirmed the identification of OSCC. And two studies were excluded as they were concerned of prognosis and only contained the cases, lacking of controls. Finally, 13 papers [20-24, 35-42] were conformed to the inclusion criteria and they were eligible for the meta-analysis of CYP1A1 Ile462Val polymorphism.
We established a database concerning of the information extracted from each included paper. Summaries of these studies were presented in Table 1 which included the first author, ethnicity, country, number and characteristics of cases and controls, and other relevant information. Of the 13 studies included in this meta-analysis, eight studies were performed in Asians, four in Caucasians, and one in mixed race. The number of cases and controls in the papers included in this meta-analysis for varied from 31 to 190, 60 to 241, respectively. The frequency of the CYP1A1 Ile462Val homozygous variant allele (Val/Val) in cases group varied from 0-13.4%, and the controls group were present in 0-6.2%.
The results of quality assessment
According to NOS system, of all observational studies are awarded a maximum of four stars in selection, two stars in comparability, and three stars in exposure. In this meta-analysis, the assessment results for the selected studies ranges from six to eight stars (Table 1), indicating all of the selected studies are moderate-high qualities in methodology.
Test of heterogeneity and quantitative synthesis
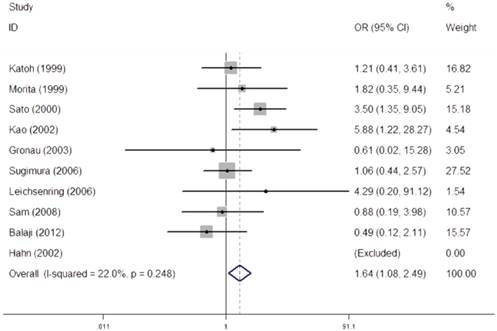
We performed a heterogeneity analysis of the dominant model (Val/Val + Val/Ile vs. Ile/Ile), recessive model (Val/Val vs. Val/Ile + Ile/Ile), homozygous model (Val/Val vs. Ile/Ile), and allele comparing (Val vs. Ile). Owing to no heterogeneity was observed in recessive model (I2=22.0%, PQ- test=0.248), fixed-effect models were used to synthesized the data (Figure 2). The overall results in recessive model suggested that individuals carrying homozygous variant (Val/Val) might have an increased risk of OSCC when compared with those who carried the homozygous Ile allele (OR=1.64, 95% CI=1.08-2.49).
Characteristics of the studies evaluating the association of CYP1A1 Ile 462 Val polymorphism with the susceptibility of OSCC
| Year | First author | Ethnicity | Country | cases | controls | Control source | Matching | NOS | HWE | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Val/Val | Val/Ile | Ile/Ile | All | Val/Val | Val/Ile | Ile/Ile | All | ||||||||
| 1997 | Park | Caucasian | USA | 23* | 108 | 131 | 10* | 121 | 131 | Healthy | Age, gender | 8 | Yes | ||
| 1999 | Katoh | Asian | Japan | 6 | 36 | 50 | 92 | 8 | 50 | 89 | 147 | Non-cancer | --- | 6 | Yes |
| 1999 | Morita | Asian | Japan | 2 | 6 | 23 | 31 | 6 | 54 | 104 | 164 | Healthy | --- | 7 | Yes |
| 2000 | Sato | Asian | Japan | 19 | 55 | 68 | 142 | 6 | 46 | 90 | 142 | Non-cancer | Age, gender | 8 | Yes |
| 2002 | Hahn | Caucasian | Germany | 0 | 4 | 90 | 94 | 0 | 6 | 86 | 92 | Healthy | --- | 6 | Yes |
| 2002 | Kao | Asian | China | 8 | 84 | 14 | 106 | 2 | 78 | 66 | 146 | Non-cancer | --- | 6 | Yes |
| 2003 | Gronau | Caucasian | Germany | 0 | 18 | 55 | 73 | 1 | 35 | 100 | 136 | Non-cancer | Age, smoking &alcohol habits | 8 | Yes |
| 2006 | Sugimura | Asian | Japan | 8 | 33 | 81 | 112 | 15 | 88 | 138 | 241 | Non-cancer | --- | 6 | Yes |
| 2006 | Leichsenring | Mixed | Brazil | 2 | 18 | 52 | 72 | 0 | 16 | 44 | 60 | Healthy | Age, gender, smoking habits | 8 | Yes |
| 2008 | Buch | Caucasian | USA | 28* | 162 | 190 | 65* | 345 | 410 | Healthy | Age, gender | 8 | NA | ||
| 2008 | Sam | Asian | India | 3 | 35 | 149 | 187 | 4 | 36 | 180 | 220 | Non-cancer | Age, gender | 8 | Yes |
| 2009 | Amtha | Asian | Indonesia | 36* | 45 | 81 | 85* | 77 | 162 | Non-cancer | Age, gender | 8 | NA | ||
| 2012 | Balaji | Asian | India | 3 | 34 | 120 | 157 | 5 | 26 | 101 | 132 | Non-cancer | Age | 6 | Yes |
NA: not available; HWE: Hardy-Weinberg Equilibrium; *: Val/Val + Val/Ile
Note: control source, Non-cancer means patients are in hospital without cancers.
Meta-analysis for the association between CYP1A1 Ile462Val polymorphism and OSCC risk under recessive model (Val/Val vs. Val/Ile + Ile/Ile).
As for the dominant model (I2=72.0%, PQ- test=0.000), homozygous model (I2=54.7%, PQ- test=0.024), random-effect models were used to synthesize the data, respectively. However, results in dominant model, homozygous model failed to obvious association of CYP1A1 Ile462Val with risks of OSCC. In order to further explore the observed heterogeneity in dominant model and additive model, subgroup analyses were performed by ethnicity, source of control, and HWE and the details were shown in Table 2.
As for allele comparing model, obvious heterogeneity were observed (I2 =66.4%; PQ- test=0.002), but the result had no statistical significance (P=0.308). The details of allele comparing model were also shown in the Table 2.
Publication bias
The Begg's funnel plot was used to evaluate the possible publication bias. Besides, the Begg's test and Egger's linear regression were used to the quantitative evaluation of the symmetry of the meta-analysis funnel plots and the results were as follows: (1) dominant model: Begg's Test, P=0.502 and Egger's linear regression test: t=0.49, P=0.634; (2) recessive model: Begg's Test, P=0.230 and Egger's linear regression test: t=0.26, P=0.803; (3) homozygous model: Begg's Test, P=0.230 and Egger's linear regression test: t=0.89, P=0.416; (4) allele comparing model: Begg's Test, P=0.371 and Egger's linear regression test: t=-1.42, P=0.192. The data of all four models indicated that there were not significant publication biases for all them.
Sensitivity analysis
In order to evaluate the stability of the results and reveal the influence of each study on the pooled ORs, sensitivity analysis were performed by excluding each case-control study respectively. When all of the four models were performed sensitivity analysis, the estimates in these four models changed between lower CI limits and upper CI limits, suggesting the results in this meta-analysis were stable.
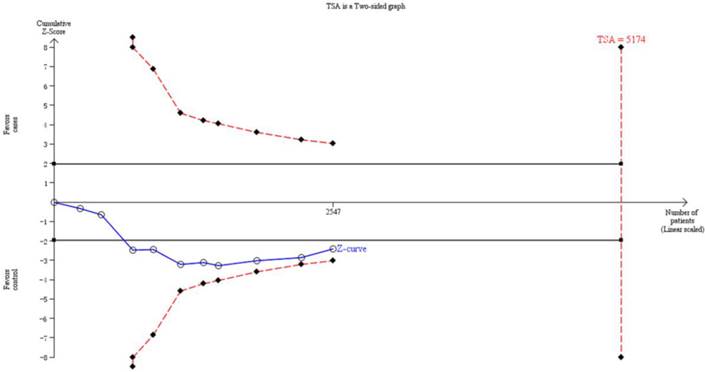
Trial sequential analysis
Thirteen trials (3651 subjects) were used to investigate the association between CYP1A1 Ile462Val polymorphism and OSCC risk. Using the recessive model (including 10 trials with 2547 subjects) as an example, we performed the TSA and found that the required information size to demonstrate clear conclusions was 5174 subjects (Figure 3). The cumulative z-curve does not cross the trial monitoring boundary before reaching the required power, which indicates that the cumulative evidence is insufficient and further trials are necessary (Figure 3). The results of other models were not shown as the study methods were similar.
Discussion
Oral cancer is the cancer of mouth, including squamous cell carcinoma, adenocarcinoma, verrucous carcinoma, and so on. Different histopathologic types of cancers might have different genetic susceptibilities, for example, CYP1A1 Ile462Val polymorphism is a risk factor of squamous cell carcinoma of lung, but the associations vary in different histological types of lung cancer [17, 43]. Therefore, it is more reasonable to assess the association of gene polymorphisms with risks of OSCC, oral adenocarcinoma and other types, separately.
Meta-analysis of the association between CYP1A1 Ile 462 Val polymorphism and OSCC risk
| Variables | Val/Val+Val/Ile vs. Ile/Ile | Val/Val vs. Val/Ile+Ile/Ile | Val/Val vs. Ile/Ile | Val vs. Ile | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR(95% CI) | I2(%) | P (Q- test) | P | OR(95% CI) | I2 (%) | P(Q- test) | P | OR(95% CI) | I2 (%) | P(Q- test) | P | OR(95% CI) | I2 (%) | P(Q- test) | P | ||||
| Total | 1.17 (0.86-1.61) | 72 | <0.001 | 0.321 | 1.64 (1.08-2.49) | 22.0 | 0.248 | 0.019 | 1.74 (0.84-3.60) | 54.7 | 0.024 | 0.133 | 1.16 (0.88-1.52) | 66.4 | 0.002 | 0.308 | |||
| Ethnicity | |||||||||||||||||||
| Caucasian | 1.12 (0.66-1.88) | 50.4 | 0.109 | 0.676 | 0.61 (0.02-15.28) | --- | --- | 0.766 | 0.60 (0.02-15.07) | --- | --- | 0.758 | 1.22 (0.87-1.70) | 0.0 | 0.652 | 0.537 | |||
| Asian | 1.21 (0.78-1.89) | 80.9 | <0.001 | 0.398 | 1.63 (1.07-2.50) | 36.9 | 0.147 | 0.024 | 1.75 (0.79-3.92) | 64.5 | 0.01 | 0.169 | 0.84 (0.49-1.45) | 75.5 | 0.0 | 0.254 | |||
| Mixed | 1.06 (0.49-2.29) | --- | --- | 0.887 | 4.29 (0.20-91.12) | --- | --- | 0.35 | 4.24 (0.20-90.62) | --- | --- | 0.355 | 1.17 (0.59-2.35) | --- | --- | 0.654 | |||
| Control Source | |||||||||||||||||||
| Healthy | 1.04 (0.65-1.67) | 46.3 | 0.114 | 0.876 | 2.38 (0.59-9.65) | 38.6 | 0.62 | 0.225 | 1.97 (0.44-8.22) | 0.0 | 0.554 | 0.386 | 0.91 (0.57-1.45) | 0.0 | 0.603 | 0.682 | |||
| Non-cancer | 1.25 (0.82-1.91) | 79.9 | 0.000 | 0.297 | 1.59 (1.03-2.46) | 0.0 | 0.134 | 0.037 | 1.69 (0.72-4.00) | 65.3 | 0.008 | 0.231 | 1.23 (0.89-1.70) | 74.9 | 0.001 | 0.214 | |||
| HWE | |||||||||||||||||||
| Yes | 1.27 (0.87-1.83) | 73.9 | 0.000 | 1.64 (1.08-2.49) | 22.0 | 0.248 | 0.019 | 1.74 (0.84-3.60) | 54.7 | 0.024 | 0.133 | 1.16 (0.88-1.52) | 66.4 | 0.002 | 0.308 | ||||
| NA | 0.83 (0.58-1.18) | 0.0 | 0.521 | --- | --- | --- | --- | --- | --- | --- | --- | --- | --- | --- | --- | ||||
I2: the variation in OR attributable to heterogeneity; P(Q-test)>0.05, heterogeneity was not statistically significant; P>0.05, no statistical significant
The information size to demonstrate the association between CYP1A1 Ile462Val polymorphism and OSCC risk. The solid line represents the cumulative Z-curve. The dashed curve represents the trial sequential monitoring boundary.
To our knowledge, this is the first meta-analysis to assess the association between CYP1A1 Ile462Val polymorphism and OSCC risk. Although there were two previous meta-analyses [44, 45] concerning of CYP1A1 Ile462Val polymorphism and risks of oral cancer, the results did not involve in single histopathologic type and therefore they failed to be on behalf of the association of CYP1A1 Ile462Val polymorphism with OSCC risk. For instance, Singh, et al. [46] found no association between CYP1A1 Ile462Val polymorphism and oral cancer risk. However, the cases included in this research are OSCC and verrucous carcinoma. Thus, the results might fail to be on behalf of the association of OSCC and CYP1A1 Ile462Val polymorphism. Based on what mentioned above and for obtaining a powerful conclusion concerning about risks of OSCC and CYP1A1 Ile462Val polymorphism, we performed this systematical meta-analysis to evaluate it.
In the present meta-analysis, for overall data, the results of recessive model showed that the homozygous Val allele mutation (Val/Val) might play a critical role in the occurrence and development of OSCC (OR=1.64, 95% CI=1.08-2.49), which not only suggested that the same homogeneity of effect size was across all relevant studies but also were inconsistent with the previous meta-analyses [44, 45]. As the results shown, the individuals who carried homozygous mutation (Val/Val) compared with those who carried wild genotype (Ile/Ile) and heterozygous variant (Val/Ile) might have an increased susceptibility of OSCC, implying that the OSCC might have different susceptibility from other histological types of oral cancer. As for the other three models (homozygous model, dominant model and allele comparing model), they failed to show obvious association between CYP1A1 Ile462Val polymorphism and OSCC risk, and the heterogeneities among studies were observed in them, respectively.
In order to explore the source of heterogeneity, we performed the subgroup analyses. After the subgroup analyses by ethnicity, source of control, HWE, the heterogeneities were not removed, indicating that other factors, such as age, gender, country, lifestyle, social status, smoking and alcohol habits might also yield to heterogeneities. The confused results may be resulted from few studies, few subjects, or other potential factors, and the more credible conclusions need the future studies containing large sample sizes and well-designed criteria to confirm.
Inevitably, several disadvantages should be noticed. First, the selected papers only involved in data of Asia, America, Europe, but without the related data about Africa and Australia. People come from different places may share different genotypes, thus, applicability of the results are limited. Second, in the current meta-analysis, only 13 articles with 1468 cases and 2183 controls were included. Therefore, the study might only have limited statistical power. Third, we performed a subgroup analysis by ethnicity, source of control, HWE, but the other factors, such as gender, age, were not performed for data limitations. Finally, heterogeneity was existed, which might weaken the reliability of conclusions. Based on the limitations mentioned above, the results should be considered with caution.
Overall, in spite of these limitations, the results of this analysis suggest that the homozygous variant might be a risk factor of OSCC. And future studies focusing on CYP1A1 Ile462Val polymorphism containing large sample sizes and well-designed criteria are necessary to make the conclusions more credible.
Abbreviations
OSCC, oral squamous cell carcinomas; HGP, human genome project; SNP, single nucleotide polymorphisms; GST, Glutathione S-transferase; CYP1A1, Cytochrome P4501 A1; AHH, aryl hydrocarbon hydrolase; PAH, polycyclic aromatic hydrocarbon; HWE, Hardy-Weinberg Equilibrium; NOS, Newcastle-Ottawa Scale; OR, odds ratio; CI, confidence interval; TSA, trial sequential analysis.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (Grant No. 81101566).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Kademani D. Oral cancer. Mayo Clin Proc. 2007;82:878-87
2. Petersen PE. Oral cancer prevention and control--the approach of the World Health Organization. Oral Oncol. 2009;45:454-60
3. Barnes L, Eveson JW, Reichart P, Sidransky D. Pathology and genetics of head and neck tumours: Classification of tumours. Lyon, France: IARC Press. 2005
4. Chen GS, Chen CH. [A study on survival rates of oral squamous cell carcinoma]. Kaohsiung J Med Sci. 1996;12:317-25
5. Lim YC, Choi EC. Surgery alone for squamous cell carcinoma of the oral cavity: survival rates, recurrence patterns, and salvage treatment. Acta Otolaryngol. 2008;128:1132-7
6. Lo Muzio L, Campisi G, Farina A, Rubini C, Pannone G, Serpico R. et al. P-cadherin expression and survival rate in oral squamous cell carcinoma: an immunohistochemical study. BMC Cancer. 2005;5:63
7. Sujatha D, Hebbar PB, Pai A. Prevalence and correlation of oral lesions among tobacco smokers, tobacco chewers, areca nut and alcohol users. Asian Pac J Cancer Prev. 2012;13:1633-7
8. Krishna Rao SV, Mejia G, Roberts-Thomson K, Logan R. Epidemiology of oral cancer in Asia in the past decade--an update (2000-2012). Asian Pac J Cancer Prev. 2013;14:5567-77
9. Blot WJ, McLaughlin JK, Winn DM, Austin DF, Greenberg RS, Preston-Martin S. et al. Smoking and drinking in relation to oral and pharyngeal cancer. Cancer Res. 1988;48:3282-7
10. Figuero Ruiz E, Carretero Pelaez MA, Cerero Lapiedra R, Esparza Gomez G, Moreno Lopez LA. Effects of the consumption of alcohol in the oral cavity: relationship with oral cancer. Med Oral. 2004;9:14-23
11. Johnson N. Tobacco use and oral cancer: a global perspective. J Dent Educ. 2001;65:328-39
12. Peng J, Liu HZ, Zhu YJ. Null Glutathione S-transferase T1 and M1 genotypes and oral cancer susceptibility in China and India--a meta-analysis. Asian Pac J Cancer Prev. 2014;15:287-90
13. Masood N, Yasmin A, Kayani MA. Genetic deletions of GSTM1 and GSTT1 in head and neck cancer: review of the literature from 2000 to 2012. Asian Pac J Cancer Prev. 2013;14:3535-9
14. Zhuo X, Zhao H, Chang A, Zhou Y, Zhang G, Song Y. et al. Quantitative assessment of CYP1A1*2A variations with oral carcinoma susceptibility: evidence from 1,438 cases and 2,086 controls. Cancer Invest. 2012;30:552-9
15. Wang Y, Yang H, Li L, Wang H. Glutathione S-transferase T1 gene deletion polymorphism and lung cancer risk in Chinese population: a meta-analysis. Cancer Epidemiol. 2010;34:593-7
16. Pei YL, Zhang HL, Han HG. Polymorphism of 8q24 rsl3281615 and breast cancer risk: a meta-analysis. Tumour Biol. 2013;34:421-8
17. Zhang Y, Wang MY, He J, Wang JC, Yang YJ, Jin L. et al. Tumor necrosis factor-alpha induced protein 8 polymorphism and risk of non-Hodgkin's lymphoma in a Chinese population: a case-control study. PLoS One. 2012;7:e37846
18. Hashibe M, Brennan P, Strange RC, Bhisey R, Cascorbi I, Lazarus P. et al. Meta- and pooled analyses of GSTM1, GSTT1, GSTP1, and CYP1A1 genotypes and risk of head and neck cancer. Cancer Epidemiol Biomarkers Prev. 2003;12:1509-17
19. Feng X, Zhou HF, Zheng BS, Shi JJ, Luo C, Qin JJ. Association of glutathione S-transferase P1 gene polymorphism with the histological types of lung cancer: a meta-analysis. Mol Biol Rep. 2013;40:2439-47
20. Sato M, Sato T, Izumo T, Amagasa T. Genetically high susceptibility to oral squamous cell carcinoma in terms of combined genotyping of CYP1A1 and GSTM1 genes. Oral Oncol. 2000;36:267-71
21. Park JY, Muscat JE, Ren Q, Schantz SP, Harwick RD, Stern JC. et al. CYP1A1 and GSTM1 polymorphisms and oral cancer risk. Cancer Epidemiol Biomarkers Prev. 1997;6:791-7
22. Kao SY, Wu CH, Lin SC, Yap SK, Chang CS, Wong YK. et al. Genetic polymorphism of cytochrome P4501A1 and susceptibility to oral squamous cell carcinoma and oral precancer lesions associated with smoking/betel use. J Oral Pathol Med. 2002;31:505-11
23. Morita S, Yano M, Tsujinaka T, Akiyama Y, Taniguchi M, Kaneko K. et al. Genetic polymorphisms of drug-metabolizing enzymes and susceptibility to head-and-neck squamous-cell carcinoma. Int J Cancer. 1999;80:685-8
24. Amtha R, Ching CS, Zain R, Razak IA, Basuki B, Roeslan BO. et al. GSTM1, GSTT1 and CYP1A1 polymorphisms and risk of oral cancer: a case-control study in Jakarta, Indonesia. Asian Pac J Cancer Prev. 2009;10:21-6
25. Bartsch H, Nair U, Risch A, Rojas M, Wikman H, Alexandrov K. Genetic polymorphism of CYP genes, alone or in combination, as a risk modifier of tobacco-related cancers. Cancer Epidemiol Biomarkers Prev. 2000;9:3-28
26. Landi MT, Bertazzi PA, Shields PG, Clark G, Lucier GW, Garte SJ. et al. Association between CYP1A1 genotype, mRNA expression and enzymatic activity in humans. Pharmacogenetics. 1994;4:242-6
27. Lukowsky A, Sterry W, Schneider-Burrus S. Prevalence of the MspI and Ile462Val SNPs of cytochrome P-450 1A1 in hidradenitis suppurativa. Exp Dermatol. 2010;19:541-2
28. Zhou SF, Liu JP, Chowbay B. Polymorphism of human cytochrome P450 enzymes and its clinical impact. Drug Metab Rev. 2009;41:89-295
29. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603-5
30. Cochran W. The combination of estimates from different experiments. Biometrics. 1954;10:101-29
31. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629-34
32. Brok J, Thorlund K, Wetterslev J, Gluud C. Apparently conclusive meta-analyses may be inconclusive--Trial sequential analysis adjustment of random error risk due to repetitive testing of accumulating data in apparently conclusive neonatal meta-analyses. Int J Epidemiol. 2009;38:287-98
33. Wetterslev J, Thorlund K, Brok J, Gluud C. Trial sequential analysis may establish when firm evidence is reached in cumulative meta-analysis. J Clin Epidemiol. 2008;61:64-75
34. Xie S, Shan XF, Shang K, Xu H, He J, Cai ZG. Relevance of LIG4 gene polymorphisms with cancer susceptibility: evidence from a meta-analysis. Sci Rep. 2014;4:6630
35. Sugimura T, Kumimoto H, Tohnai I, Fukui T, Matsuo K, Tsurusako S. et al. Gene-environment interaction involved in oral carcinogenesis: molecular epidemiological study for metabolic and DNA repair gene polymorphisms. J Oral Pathol Med. 2006;35:11-8
36. Lakkakula V.K.S. BhaskarSam SS, Thomas V, Reddy SK, Surianarayanan G, Chandrasekaran A. CYP1A1 polymorphisms and the risk of upper aerodigestive tract cancers in an Indian population. Head Neck. 2008;30:1566-74
37. Leichsenring A, Losi-Guembarovski R, Maciel ME, Losi-Guembarovski A, Oliveira BW, Ramos G. et al. CYP1A1 and GSTP1 polymorphisms in an oral cancer case-control study. Braz J Med Biol Res. 2006;39:1569-74
38. Katoh T, Kaneko S, Kohshi K, Munaka M, Kitagawa K, Kunugita N. et al. Genetic polymorphisms of tobacco- and alcohol-related metabolizing enzymes and oral cavity cancer. Int J Cancer. 1999;83:606-9
39. Hahn M, Hagedorn G, Kuhlisch E, Schackert HK, Eckelt U. Genetic polymorphisms of drug-metabolizing enzymes and susceptibility to oral cavity cancer. Oral Oncol. 2002;38:486-90
40. Gronau S, Koenig-Greger D, Jerg M, Riechelmann H. GSTM1 enzyme concentration and enzyme activity in correlation to the genotype of detoxification enzymes in squamous cell carcinoma of the oral cavity. Oral Dis. 2003;9:62-7
41. Buch SC, Nazar-Stewart V, Weissfeld JL, Romkes M. Case-control study of oral and oropharyngeal cancer in whites and genetic variation in eight metabolic enzymes. Head Neck. 2008;30:1139-47
42. Balaji L, Singh KB, Bhaskar LV. CYP1A1 genotypes and haplotypes and risk of oral cancer: A case-control study in South Indians. Genet Mol Biol. 2012;35:407-12
43. Zhan P, Wang Q, Qian Q, Wei SZ, Yu LK. CYP1A1 MspI and exon7 gene polymorphisms and lung cancer risk: an updated meta-analysis and review. J Exp Clin Cancer Res. 2011;30:99
44. Zhuo X, Zhao H, Chang A, Ye H, Zhou Y, Song Y. et al. Cytochrome P450 1A1 Ile462Val polymorphism and oral carcinoma risk: an updated meta-analysis including 1,515 cases and 2,233 controls. Tumour Biol. 2012;33:2079-89
45. Zhuo WL, Wang Y, Zhuo XL, Zhu Y, Wang WD, Zhu B. et al. CYP1A1 and GSTM1 Polymorphisms and Oral Cancer Risk: Association Studies Via Evidence-Based Meta-Analyses. Cancer Invest. 2009;27:86-95
46. Singh R, Haridas N, Shah F, Patel J, Shukla S, Patel P. Gene polymorphisms, tobacco exposure and oral cancer susceptibility: a study from Gujarat, West India. Oral Dis. 2014;20:84-93
Author contact
![]() Corresponding author: Su-Xiu Chen, Department of General Medicine, The First Affiliated Hospital of Wenzhou Medical University, No.2 Fuxue Lane, Wenzhou 325000, Zhejiang, China. Tel: (+86-0577) 88069359, Fax: (+86-0577) 88069372, E-mail: chensuxiu100com; or Zhi-Gang Cai, Department of Oral and Maxillofacial Surgery, Peking University School and hospital of Stomatology, 22# Zhongguancun South Avenue, Beijing 100081, China. Tel: (+86-010) 82195956, Fax: (+86-010) 62173402; E-mail: c2013xscom.
Corresponding author: Su-Xiu Chen, Department of General Medicine, The First Affiliated Hospital of Wenzhou Medical University, No.2 Fuxue Lane, Wenzhou 325000, Zhejiang, China. Tel: (+86-0577) 88069359, Fax: (+86-0577) 88069372, E-mail: chensuxiu100com; or Zhi-Gang Cai, Department of Oral and Maxillofacial Surgery, Peking University School and hospital of Stomatology, 22# Zhongguancun South Avenue, Beijing 100081, China. Tel: (+86-010) 82195956, Fax: (+86-010) 62173402; E-mail: c2013xscom.

 Global reach, higher impact
Global reach, higher impact