Impact Factor
ISSN: 1837-9664
J Cancer 2014; 5(9):761-764. doi:10.7150/jca.10360 This issue Cite
Short Research Communication
CD70: A Potential Target in Breast Cancer?
1. Department of Medical Oncology, Centre Henri Becquerel, Rouen, France;
2. Department of Pathology, Centre Henri Becquerel, Rouen, France;
3. INSERM U918, Centre Henri Becquerel, IRIB, Rouen, France.
Received 2014-8-18; Accepted 2014-9-29; Published 2014-10-22
Abstract
CD70 is a co-stimulatory molecule involved in the immune response and also in cancer development and progression. Recent studies show that high CD70 expression in cancer cells may inhibit the anti-tumor response. Furthermore, CD70 expression has been reported as a predictive marker of resistance to chemotherapy in ovarian cancers. Some in vitro studies have shown that CD70 expression is epigenetically down-regulated through hypermethylation of its promoter during tumoral progression. This study evaluated the level of CD70 expression in surgical samples of breast invasive tumors and determined its correlation with CD70 promoter methylation.
Twenty “luminal A” and 20 “basal-like” frozen samples from early breast tumors were retrospectively selected. CD70 expression was evaluated by quantitative real-time PCR. Total DNA was bisulfite-treated, and methylation levels of 5 consecutive CG sites present in the proximal region (-464, -421) of the promoter were assessed by pyrosequencing analysis. Statistical analyses were performed using the Mann-Whitney test.
The median relative CD70 expression level was 0.37 and was significantly higher in the basal-like group (0.78 [0.24-31.7]) compared to the luminal A group (0.25 [0.03-1.83], p=0.0001). The median methylation level was 61%, with no significant difference between the basal-like (63%) and luminal A (58%) groups. No correlation was found between CD70 expression and CD70 methylation level.
In this study, higher CD70 expression was observed in the basal-like group, but this expression was not related to promoter methylation. The higher expression in the poor-prognosis subgroup of patients makes CD70 a potential target for emerging anti-CD70 therapies.
Keywords: breast cancer, CD70, DNA methylation, mRNA expression, q-RT-PCR, antibody-drug conjugate.
Introduction
Tumor cells develop many mechanisms to escape the immune system, including the loss of tumor surface antigen expression, induction of lymphocytes apoptosis, and inhibition of some co-stimulatory molecules involved in recognition by lymphocytes [1].
CD70 (encoded by the TNFSF7 (Tumor Necrosis Factor ligand Superfamily member 7) gene) is a co-stimulatory factor present on B-cells, activated T-cells, and dendritic cells [2]. The interaction with its ligand, CD27, is involved in survival, proliferation, and lymphocyte differentiation [3]. CD70 is not expressed in non-lymphoid normal tissue [4] but is overexpressed in tumor cells of various solid cancers [5]. This high expression may induce cytotoxic effects in B-cells and T-cells, induce lymphocyte apoptosis, and promote escape from the immune system [6]. In contrast, Aulwurm and al. [7] suggested that CD70 expression could induce a long-lasting immunity in mice models of gliomas. Furthermore, many in vitro studies have shown that CD70 overexpression by tumor cells can induce an anti-tumor response via T-cells in breast cancer cell lines [8,9]. In breast cancer, the role of CD70 also remains unclear. Rhee and al. showed that its expression in breast cancer cell lines is inhibited during tumor cell progression, most likely via promoter hypermethylation [10]. One of the hypotheses is that the inhibition of CD70 expression in tumor cells would allow them to escape the immune response. The study of the CD70 promoter region (which contains 3 CpG islands) was originally described by Lu et al.[11] focusing on a 1-kb sequence located upstream of the transcription initiation site and containing the second CpG island. Yu et al.[12] then showed that this second CpG island is heavily methylated during the in vitro proliferation of mammary tumor cells. This observation was recently confirmed by a study showing that CD70 gene expression is inhibited via the hypermethylation of its promoter in a breast cancer cell line [10]. To our knowledge, no study to date has been conducted in vivo to evaluate the expression and methylation of CD70 in breast cancer. Since "luminal A" and "basal-like" tumors present opposed biological and clinical behaviours, we hypothesised that their relationship with immunity may differ.
Accordingly, the aim of this study was to evaluate the level of CD70 expression in surgical samples of 2 groups of breast invasive tumors, basal-like and luminal A, and to determine the correlation of the expression with its promoter methylation.
Materials and Methods
Patients and Samples
Fifty-six patients aged ≥18 years who had been treated for newly diagnosed early breast cancer in Henri Becquerel Center, Rouen, France, between January 2008 and December 2011 were retrospectively screened. Invasive breast carcinomas that were treated first by surgery without neo-adjuvant treatments were eligible. Twenty tumors with a “luminal A” immunohistochemical profile and twenty tumors with a “basal-like” profile were selected. The “luminal A” tumors were positive for Estrogen Receptors (ER) (≥10%) and negative for HER2 overexpression/amplification and showed a low Ki67 (≤20%). The “basal-like” tumors were negative for ER (<10%), negative for Progesterone Receptors (PR) and negative for HER2 overexpression/amplification, showed high Ki67 [13] (>20%), and were positive for at least one basal-cell marker (Cytokeratin 5 or EGF-R)[14]. For each patient, informed consent for biomedical studies on histological samples was collected. Tumor DNA and RNA were extracted from frozen tissue using the AllPrep DNA/RNA Mini Kit (QIAGEN).
CD70 expression
Quantitative Real-Time RT-PCR was performed using predesigned TaqMan® Gene Expression Assays (Life Technologies). Analyses were performed using a 7900HT Fast Real Time PCR System (Life technologies). The relative expression level of target genes within a sample was expressed using the DCt method [15]. Ct is the number of cycles of amplification needed to reach a predefined threshold and DCt is the difference between the Ct for the gene of interest and the Ct for the reference gene (HMBS). DCt is proportional to the amount of RNA in the sample and allows the evaluation of the relative expression level for each sample. DCt values were averaged for duplicates.
CD70 promoter methylation
The DNA samples were adjusted to a concentration of 100 ng/μl and then treated with bisulfite using an EZ DNA Methylation-Gold Kit (ZYMO RESEARCH). The target region (the second CpG island of the promoter region) was then amplified by two consecutive PCR (conventional PCR and nested PCR, according to the technique used in lymphomas by Bertrand et al. [16]). In this study, CG pairs 6-10 were studied. Genomic localizations (genome release GRCh37) of CG sites 6, 7, 8, 9, and 10 were 6 591 628, 6 591 610, 6 591 602, 6 591 599, and 6 591 586, respectively. The analysis was performed by pyrosequencing using a PyroMark® Q24 (Qiagen).
Statistical analysis
Statistical analyses were performed with the MedCalc® software. Comparisons of CD70 expression and levels of methylation between the two groups of tumors were performed using the non-parametric Mann-Whitney U test. The correlation between methylation and expression was studied by the Spearman coefficient.
Results
Patients and Samples
Among the 40 patients, the median age was 60 (from 30 to 88 years old). Twenty patients (50%) had lymph node involvement at diagnosis. The median tumor size was 28 mm (20-150 mm). A histological analysis revealed ductal invasive carcinoma in 38 cases, mucinous carcinoma in 1 case, and metaplastic squamous cell carcinoma in 1 case. The median SBR grade was III. Twenty tumors (50%) significantly expressed ER, and 14 tumors (35%) significantly expressed PR. No tumor overexpressed HER2. Ki67 was ≥20% in 19 of the 40 tumors (47%). Of the basal-like tumors, CK5 was positive in 18 of 20 cases (90%), and EGF-R was positive in 7 of 20 cases (35%); all basal-like tumors were positive for at least one of the two markers. The patient characteristics according to their group are detailed in Table 1.
CD70 expression
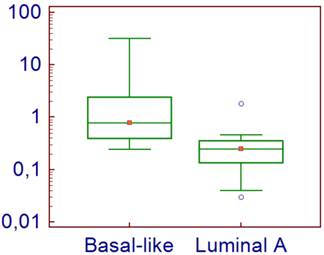
For the 40 analyzed tumors, the normalized level of expression of CD70 ranged from 0.03 to 31.70, with a median of 0.37. The expression level was significantly higher for the basal-like tumors (median = 0.78 [0.24-31.7]) compared with the luminal A tumors (median = 0.25 [0.03-1.83]) (p=0.0001) (Figure 1).
Patients and tumors characteristics.
| Luminal A | Basal-like | |
|---|---|---|
| Median age (years) | 64 | 56 |
| Median tumor size (mm) | 27 | 29 |
| Median grade (SBR) | II | III |
| Node positive (n=) | 11 (55%) | 9 (45%) |
| Histological type (n=) | ||
| Ductal invasive | 19 (95%) | 19 (95%) |
| Other type | 1 (5%) | 1 (5%) |
| ER ≥ 10% (n=) | 20 (100%) | 0 (0%) |
| PR ≥ 10% (n=) | 14 (70%) | 0 (0%) |
| HER2 overexpression/amplification | 0 (0%) | 0 (0%) |
| Ki67<10% (n=) | 13 (65%) | 1 (5%) |
| 10%≤Ki67<20% (n=) | 7 (35%) | 0 (0%) |
| Ki67≥20% | 0 (0%) | 19 (95%) |
| Median Ki67 | 10% | 60% |
CD70 Expression levels in tumor cells according to their subtype.
CD70 promoter methylation
The overall median methylation level of the 5 studied CG sites was 64% (13-92%): 65% in the basal-like and 58% in the luminal A group. A trend toward a greater methylation of CG #1 in the basal-like tumors compared with the luminal A tumors was observed (56.5% vs 43%, respectively, p=0.07). There was no significant difference in methylation levels between the 2 groups for the 4 other studied CG sites (#2: 69% vs 59%, #3: 81% vs 82%, #4: 61.5% vs 54% and #5: 65.5% vs 59.5% for basal like and luminal A, respectively).
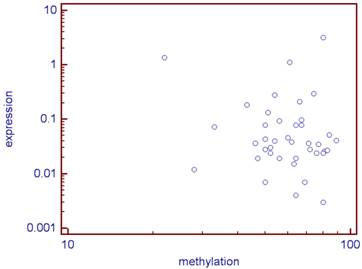
There was no statistical link between the methylation levels of the 5 studied CG sites and the expression of CD70 (correlation coefficient: r =-0.09, p=0.56; Figure 2).
Correlation between the CD70 expression and its median methylation rate of the five studied CG (log) for the 40 analyzed tumors.
Discussion
Few authors have studied the expression and methylation of CD70 in breast cancer, particularly in ex vivo tumors. Yu and al.[12] studied the methylation of the CD70 promoter in breast cancer cell lines and found that the proximal promoter region of CD70 was hypermethylated at the time of tumor progression, in parallel with a very low RNA expression. However, we did not confirm these results, and the difference in results may be related to the use of a different technique for the assessment of methylation level (cloning versus pyrosequencing) and tumor cells of different origin (cell lines versus ex vivo). Although the expression of CD70 was significantly higher in the basal-like group, we did not observe lower methylation levels in this group. Nevertheless, the epigenetic regulation of CD70 is currently still unknown, and many other factors may be involved in its regulation (e.g., histone modifications, transcription and transduction factors).
CD70 is currently considered to be a potentially attractive therapeutic target, because it is overexpressed in many types of lymphomas and invasive carcinomas but is not expressed in normal tissue. Indeed, several agents specifically targeting CD70 are currently being studied [17, 18].
In this study dedicated to ex vivo early breast cancer, we observed a significantly higher expression of CD70 in the group of basal-like tumors compared to luminal A tumors. Basal-like tumors are known to be more aggressive than luminal A tumors. Our results are concordant with those of Jilaveanu et al.[4], who showed that the expression of CD70 is higher in renal tumors with a poor prognosis and was associated with lower survival in clear cell carcinomas. Similarly, Bertrand showed in 2013 that the overexpression of CD70 is correlated with poorer survival [16] in diffuse large B-cell lymphoma. But to demonstrate the specific role of increased CD70 expression in basal-like breast cancer as a functional marker rather than general upregulation of stimulatory signals as a result of higher immune activation, expression levels of other co-stimulatory molecules such as CD86, CD80, CD70 together with effector cytokines should be also analyzed and compared between basal-like tumors and luminal A tumors.
Interestingly, the expression of CD70 could also be considered as a potential predictor of response to chemotherapy. Indeed, Liu and al.[19] in 2013 focused on 92 patients with stage III or IV ovarian cancer. In this group of patients, CD70 overexpression in tumor cells by immunohistochemistry was associated with a decreased response to cisplatin and increased chemoresistance, as well as a decreased survival rate.
Furthermore, the tumor stage should be considered: in metastatic situations, the overall inflammation may change the response to anti-CD70 targeted therapies.
Finally, our findings regarding differential CD70 expression in ex vivo early breast cancer between basal-like and luminal A tumors are encouraging and deserve to be confirmed using a larger cohort to assess its predictive or prognostic value and to correlate the expression of RNA with immunohistochemically detected protein expression for routine use.
Abbreviations
TNFSF7: Tumor Necrosis Factor ligand SuperFamily member 7; ER: Estrogen Receptors; PR: Progesterone Receptors.
Acknowledgements
The authors wish to thank all members of the department of molecular biology for their technical assistance and comments.
Competing Interests
The authors have not received any financial support for this study and have no conflicts of interest to declare.
References
1. Pardoll DM. Cancer vaccines. Immunol Today. 1993;14(6):310-316
2. Bowman MR, Crimmins MA, Yetz-Aldape J, Kriz R, Kelleher K, Herrmann S. The cloning of CD70 and its identification as the ligand for CD27. J Immunol Baltim Md 1950. 1994;152(4):1756-1761
3. Grewal IS. CD70 as a therapeutic target in human malignancies. Expert Opin Ther Targets. 2008;12(3):341-351
4. Jilaveanu LB, Sznol J, Aziz SA, Duchen D, Kluger HM, Camp RL. CD70 expression patterns in renal cell carcinoma. Hum Pathol. 2012;43(9):1394-1399
5. Ryan MC, Kostner H, Gordon KA, Duniho S, Sutherland MK, Yu C. et al. Targeting pancreatic and ovarian carcinomas using the auristatin-based anti-CD70 antibody-drug conjugate SGN-75. Br J Cancer. 2010;103(5):676-684
6. Wischhusen J, Jung G, Radovanovic I, Beier C, Steinbach JP, Rimner A. et al. Identification of CD70-mediated apoptosis of immune effector cells as a novel immune escape pathway of human glioblastoma. Cancer Res. 2002;62(9):2592-2599
7. Aulwurm S, Wischhusen J, Friese M, Borst J, Weller M. Immune stimulatory effects of CD70 override CD70-mediated immune cell apoptosis in rodent glioma models and confer long-lasting antiglioma immunity in vivo. Int J Cancer J Int Cancer. 2006;118(7):1728-1735
8. Cormary C, Gonzalez R, Faye J-C, Favre G, Tilkin-Mariamé A-F. Induction of T-cell antitumor immunity and protection against tumor growth by secretion of soluble human CD70 molecules. Cancer Gene Ther. 2004;11(7):497-507
9. Douin-Echinard V, Péron J-M, Lauwers-Cancès V, Favre G, Couderc B. Involvement of CD70 and CD80 intracytoplasmic domains in the co-stimulatory signal required to provide an antitumor immune response. Int Immunol. 2003;15(3):359-372
10. Rhee DK, Park SH, Jang YK. Molecular signatures associated with transformation and progression to breast cancer in the isogenic MCF10 model. Genomics. 2008;92(6):419-428
11. Lu Q, Wu A, Richardson BC. Demethylation of the same promoter sequence increases CD70 expression in lupus T cells and T cells treated with lupus-inducing drugs. J Immunol Baltim Md 1950. 2005;174(10):6212-6219
12. Yu SE, Park SH, Jang YK. Epigenetic silencing of TNFSF7 (CD70) by DNA methylation during progression to breast cancer. Mol Cells. 2010;29(2):217-221
13. Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thürlimann B, Senn H-J. et al. Strategies for subtypes--dealing with the diversity of breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011Aug;22(8):1736-47
14. Cheang MCU, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK. et al. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res Off J Am Assoc Cancer Res. 2008;14(5):1368-1376
15. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods San Diego Calif. 2001;25(4):402-408
16. Bertrand P, Maingonnat C, Penther D, Guney S, Ruminy P, Picquenot JM. et al. The costimulatory molecule CD70 is regulated by distinct molecular mechanisms and is associated with overall survival in diffuse large B-cell lymphoma. Genes Chromosomes Cancer. 2013;52(8):764-774
17. Oflazoglu E, Stone IJ, Gordon K, Wood CG, Repasky EA, Grewal IS. et al. Potent anticarcinoma activity of the humanized anti-CD70 antibody h1F6 conjugated to the tubulin inhibitor auristatin via an uncleavable linker. Clin Cancer Res Off J Am Assoc Cancer Res. 2008;14(19):6171-6180
18. Law C-L, Gordon KA, Toki BE, Yamane AK, Hering MA, Cerveny CG. et al. Lymphocyte activation antigen CD70 expressed by renal cell carcinoma is a potential therapeutic target for anti-CD70 antibody-drug conjugates. Cancer Res. 2006;66(4):2328-2337
19. Liu N, Sheng X, Liu Y, Zhang X, Yu J. Increased CD70 expression is associated with clinical resistance to cisplatin-based chemotherapy and poor survival in advanced ovarian carcinomas. OncoTargets Ther. 2013;6:615-619
Author contact
![]() Corresponding author: Camille Petrau, Department of Medical Oncology, Centre Henri Becquerel, 1 rue d'Amiens, 76038 Rouen cedex, France. Tel: (33) 232 082 222 Fax: (33) 232 082 501 E-mail: camillepetraufr.
Corresponding author: Camille Petrau, Department of Medical Oncology, Centre Henri Becquerel, 1 rue d'Amiens, 76038 Rouen cedex, France. Tel: (33) 232 082 222 Fax: (33) 232 082 501 E-mail: camillepetraufr.

 Global reach, higher impact
Global reach, higher impact