3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2014; 5(2):156-165. doi:10.7150/jca.7668 This issue Cite
Research Paper
A Majority of Low (1-10%) ER Positive Breast Cancers Behave Like Hormone Receptor Negative Tumors
1. Division of Molecular Medicine, St. John's Research Institute, Bangalore, India
2. Department of Surgical Oncology, St. John's Medical College and Hospital, Bangalore, India
3. Department of Medicine, McGill University, Montreal, Quebec, Canada, Department of Epidemiology, Biostatistics and Occupational Health, McGill University, Montreal, Quebec, Canada
4. Department of Pathology, St. John's Medical College and Hospital, Bangalore, India
5. Rangadore Memorial Hopsital, Bangalore, India
6. Shankara Cancer Hospital and Research Centre, Bangalore, India
Received 2013-9-14; Accepted 2013-12-9; Published 2014-1-23
Abstract
Background: The 2010 guidelines by ASCO-CAP have mandated that breast cancer specimens with ≥1% positively staining cells by immunohistochemistry should be considered Estrogen Receptor (ER) positive. This has led to a subclass of low-ER positive (1-10%) breast cancers. We have examined the biology and clinical behavior of these low ER staining tumors.
Methods: We have developed a probabilistic score of the “ER-positivity” by quantitative estimation of ER related gene transcripts from FFPE specimens. Immunohistochemistry for ER was done on 240 surgically excised tumors of primary breast cancer. Relative transcript abundance of 3 house-keeping genes and 6 ER related genes were determined by q-RT PCR. A logistic regression model using 3 ER associated genes provided the best probability function, and a cut-off value was derived by ROC analysis. 144 high ER (>10%), 75 ER negative and 21 low-ER (1-10%) tumors were evaluated using the probability score and the disease specific survival was compared.
Results: Half of the low-ER positive tumors were assigned to the ER negative group based on the probability score; in contrast 95% of ER negative and 92% of the high ER positive tumors were assigned to the appropriate ER group (p<0.0001). The survival of the low-ER group was intermediate between that of the high ER positive and ER negative groups (p<0.05).
Conclusion: Our results suggest that the newly lowered ASCO-CAP criteria for ER positivity, leads to the false categorization of biologically ER negative tumors as ER positive ones. This may have particular relevance to India, where we have a much higher proportion of ER negative tumors in general.
Keywords: ER 1-10 %, gene expression, q-RT-PCR, FFPE, Breast Cancer
Introduction
Estrogen receptor (ER) is an important prognostic and predictive marker in human breast cancer. Patients with tumors that are positive for ER are known to respond to endocrine therapy and have improved disease specific survival and overall survival compared to those with tumors that are ER negative [1, 2].
The technique for the estimation of ER protein has changed over time from the use of ligand binding assays, to immunohistochemical (IHC) methods of staining tumors using monoclonal antibodies of high specificity and affinity[3-5]. Over the last decade the identification of ESR1 transcripts has been shown to be a reliable method of establishing ER positivity in tumor specimens [6, 7]. In routine clinical practice IHC is still the gold standard and the level of ER is expressed as a product of the percentage of epithelial cells stained and the intensity of staining, as in the Allred score [3]. Historically the cut-off value for ER positivity was taken to be nuclear staining of 10% or more of the epithelial component of the tumor. The definitive study that confirmed the clinical utility of ER estimation by IHC to predicit responsiveness to endocrine therapy [8] comprised patients with the full spectrum of ER positivity ranging from an Allred score of 3 thru to 8. Tumors with even 1-10% of cells staining weakly demonstrated objective clinical benefit from treatment with tamoxifen [8]. In what could be construed to be an attempt to make the benifit of anti-estrogen therapy available to the widest range of patients, the American Society of Clinical Oncology (ASCO) lowered the IHC cut-off for determining ER positivity from the previous value of 10% to 1% of stained cells [9].
Perou et. al., [10] were the first to define a set of genes that were expressed at high levels in ER positive tumors compared to ER negative tumors. In addition, this study proposed the presence of a set of "intrinsic genes" that could be used to sub-classify breast tumors into four distinct sub-types; Luminal-A, Luminal-B, HER2 Enriched and Basal-like. A large number of subsequent studies have confirmed that ER positive tumors show elevated expression of "Luminal" genes that in addition to ESR1 and PgR include GATA3, TFF1, XBP1, FOXA1 amongst many others [11-14]. The early use of the expression levels of a sub-set of genes to guide clinical decision making was pioneered by the developers of Oncotype DX and MammaPrint [15, 16]. These and other studies have revealed a very high concordance of the expression of ESR1 and a restricted set of luminal genes to the level of ER protein detected by IHC [17].
While the ASCO 2010 criteria has had the desired effect of making endocrine therapy available to a greater proportion of patients it has raised uncomfortable questions about the assays and cut-offs used for the identification of ER positive tumors, and in this context the true biological nature of the low (1-10%) ER positive tumors [18, 19]. Initial studies by Iwamoto et. al.[20], and Deyarmin et. al. [21], have confirmed the concerns that a significant proportion of these low-ER positive tumors are non-luminal tumors. Indian women are known to have a greater proportion of ER negative tumors compared to Caucasian women and this trend persists even when corrected for age [22-27]. In this report we have identified women who had low-ER positive tumors and classified these tumors as either ER positive or ER negative by using cut-off values on a probabilistic logistic regression model that was based on the expression levels of a restricted set of luminal genes.
Materials and Methods
Sample and data collection
Samples for this study were obtained from our ongoing observational prospective cohort study of breast cancer patients from two tertiary care hospitals in Bangalore, India. This study was approved by the ethical review boards of the hospital involved. Informed consent for use of the specimens for research was obtained from all patients. Tissue samples from 235 untreated primary invasive breast cancer patients are included in the current study. Of the 235 patients, 5 patients had bilateral disease and both tumors were included taking the total analyzed samples to 240. These patients are part of a larger cohort of more than 450 patients who were recruited at diagnosis over a 5 year period, between June 2008 and Feb 2013. These patients are being actively followed up by a dedicated breast cancer support group “Aadhara”. We have stayed in touch with all the patients right from the time of initial diagnosis through the active treatment phase and beyond. In the case of patients who live in geographically distant sites, continuity of follow-up has been maintained through mobile-telephone contact. We have accomplished an extraordinary rate of 99% follow-up (manuscript under preparation). All clinical and histopathological details have been collected from the medical-records and updated at regular intervals. Radiologically recorded distant metastases or histologically confirmed local recurrence and death related to disease were considered as events and recorded accordingly. Sections from the surgically excised tumor tissue blocks processed in neutral-buffered formalin and paraffin-embedded (FFPE) were used for analysis. Sections, stained with hematoxylin and eosin, were used to assess the percentage of tumor epithelial cells. Only blocks with greater than 50% cancer epithelial cells were used for the analysis.
Immunohistochemistry
Our lab has been certified by the United Kingdom-National External Quality Assessment Scheme (UK-NEQAS) by participation in their assessment programme for ER IHC in 2012. IHC was done for ER and PgR according to standard procedures. Briefly, sections (5µm in thickness) were cut from FFPE blocks on the poly L-lysine (PLL) coated slides and subjected to deparaffinization in xylene and rehydrated in graded alcohol. After blocking endogenous peroxidase with a 3% hydrogen peroxide solution, antigen retrieval was done in 0.01M EDTA buffer at pH 8, in a microwave at 800 W for 2 min, 480 W for 7 min followed by 160 W for 11 min. Primary blocking was done with 1% bovine serum albumin (BSA, Sigma) for 30 min at room temperature. Primary antibodies for ER (Clone EP1, DAKO- Cat # ISO84), PgR (Clone PgR-636, DAKO, Cat # ISO68) were applied for 1 hr at room temperature. After washing with tris buffered saline with Tween (TBST--50mM/L Tris with 0.1% Tween 20), sections were further incubated with secondary antibody (DAKO REALTM EnVision TM) for 20 min as per the kit instructions. After a final wash with TBST they were developed using DAB (DAKO REALTM EnVision TM) for 10 min. Sections were counterstained with hematoxylin and mounted after dehydration in graded alcohol and xylene. Appropriate positive and negative controls were run for each batch.
Fluorescent in situ hybridization (FISH) for HER2 gene amplification
IHC results for HER2 were obtained from hospital records and all cases that were reported as positive or equivocal were further tested by FISH for HER2 amplification. Briefly, sections 5µm in thickness were cut on to PLL coated slides and incubated at 56ºC overnight. Slides were then deparaffinized in two changes of xylene for 30 min each followed by dehydration in graded alcohol. Slides were then denatured in 2X saline-sodium citrate (SSC) at 75ºC for 20 min followed by digestion with proteinase K (0.01 µg in 10% sodium lauryl sulphate) for 10 min for optimum digestion. Subsequently, slides were incubated with 4µl of HER2 probe (HER2/Neu labeled with PlatinumBright 550 and SE 17 control DNA probe labeled with PlatinumBright 495, Poseidon probes, Kreatech Diagnostics, Netherlands) in a humidified chamber for 17 hrs. Post hybridization wash was done in 2X SSC with 0.3% NP-40 solution at 75ºC for 3-4 min followed by a second wash in 2X SSC for 2 min at room temperature. Slides were further dehydrated, nucleus counterstained with DAPI, and viewed through a Olympus BX 51 fluorescent microscope for HER2 and SE 17 signals using the appropriate filter sets. Positive and negative controls were included in each batch for assay control and reproducibility.
Scoring of immuno-histochemistry and FISH
The intensity and pattern of staining were evaluated microscopically by a pathologist and recorded accordingly. Nuclear staining in >1% of the tumor cell nuclei was considered positive according to ASCO guidelines [9].
In the FISH assay, scoring of the number of signals for HER2 and SE 17 was done in at least 50 tumor cell nuclei and the ratio of HER2/SE17 signal was then calculated. Ratios above 2.2 were taken as amplified.
RNA extraction and reverse transcription
The methods used for nucleic acid extraction, quantitative RT-PCR, selection of house keeping genes (HKG) and quality control criteria for inclusion of samples in this analysis have been described in detail in a recent publication from our lab as well as in a prior report [28, 29]. In brief, total ribonucleic acid (RNA) was extracted from two 20µm sections from each patient's tumor block and the sections were deparaffinized by heat, and then subjected to overnight digestion using proteinase K (Qiagen #19133). Total RNA was then extracted using the TRI Reagent protocol according to manufacturer instructions (Sigma Aldrich # T9424). Quantitation of the RNA was done using the Ribogreen dye (Invitrogen # R11490- Quant -iT Ribogreen RNA assay kit) on a fluorescent plate reader (Tecan M200-Pro Infinite Series). 500ng of total RNA was then reverse transcribed using the ABI high capacity cDNA archive kit (ABI # 4322171) as per the manufacturer's protocol.
q-RT-PCR for the selected genes
The expression level of selected set of ER regulated genes (ESR1, PgR, GATA3, TFF1, FOXA1, XBP1) was determined along with a panel of 3 reference genes (PUM1, RPLP0, ACTβ). Primers for all genes were designed using Primer3Plus and manufactured by Eurofins, Bangalore, India. The reference genes normalize for any variations that may be introduced through variations in sample processing and handling methods which in turn lead to varied levels of RNA preservation in the FFPE blocks. The primer sequences for the genes tested are given in Table 1. Using 5ng cDNA template per reaction real time PCR was done in duplicate using SYBR Green on the LightCycler 480 II (Roche Diagnostics). Total RNA from normal human mammary gland (Clontech, USA, # 636576) and Universal Human Reference RNA (Agilent, # 740000) was also reverse transcribed and 0.1ng of this template was run in the assay as a control. Total reaction volume was 10µl. Pre-incubation and initial denaturation of the template cDNA was performed at 95˚C for 10 min, followed by amplification for 45 cycles at 95˚C for 15 sec and 60˚C for 1 min. All samples which had average Ct value of three housekeeping genes more than 32.25 were considered to have poor quality of RNA and were dropped from further analysis. Ct values for the test gene, were in turn normalized relative to the mean Ct value of the three reference genes for each sample as ∆Ct. The Relative Normalized Units (RNU) of expression of the test genes was calculated as 15- ∆Ct.
Primer sequence of the genes
| Gene | Primer Sequence | Product size |
|---|---|---|
| ACTB | F-TTCCTGGGCATGGAGTC | 85 |
| R-CAGGTCTTTGCGGATGTC | ||
| PUM1 | F-CCGGAGATTGCTGGACATATAA | 77 |
| R-TGGCACGCTCCAGTTTC | ||
| RPLPO | F-GGCTGTGGTGCTGATGGGCAAGAA | 96 |
| R-TTCCCCCGGATATGAGGCAGCAGT | ||
| ESR1 | F-GCAGGGAGAGGAGTTTGT | 65 |
| R-GACTTCAGGGTGCTGGAC | ||
| PGR | F-GACTGAGCTGAAGGCAAAGG | 76 |
| R-CGAAACTTCAGGCAAGGTGT | ||
| GATA3 | F-ATGGAGGTGACGGCGGACCA | 106 |
| R-ATGTAGGAGTGGCTGAGGCCCG | ||
| TFF 1 | F-TGCCTGCATCCTGACGCGGT | 89 |
| R-AGCGTGTCTGAGGTGTCCGGTG | ||
| XBP1 | F-GCCCAGTTGTCACCCCTCCA | 105 |
| R-GGGTCCAAGTTGTCCAGAATGCCC | ||
| FOXA1 | F-GCTACTACGCAGACACG | 69 |
| R-CTGAGTTCATGTTGCTGACC |
Statistical Analysis
Descriptive statistics of demographic and clinical variables were obtained for the whole cohort and also within sub-groups defined by the ER-IHC score. A descriptive analysis of the association between each of the genes of interest and ER positivity (as defined by ER IHC >1%) was carried out using receiver operating charactersitics (ROC) curves. To determine if a composite score of gene expression based on multiple genes could predict the ER positivity, we fit several binomial logistic regression models with ER status by IHC (>1%) as outcome and different combinations of test genes as the potential predictors. In these models, each gene receives a weight in the model that is determined by the regression coefficient. The different models were compared using the Bayesian Information Criterion (BIC). The predicted probability from the best fitting model (with lowest BIC) was compared in three groups determined by the ER IHC score (<1%, 1-10% and >10%). The ROC curve corresponding to the best fitting model was plotted, and the cut-off on the probability score corresponding to the point where both sensitivity and specificity were maximized was selected to classify patients into ER positive and ER negative categories. Kaplan-Meier survival curves and log rank tests were used to compare the survival between the three IHC groups as well as that between the groups defined by ER positive and negative as per the probability score obtained from by the logistic regression model.
Results
Table 2 shows the demographic and clinico-pathological characteristics of the patients in our cohort. The median age of the entire cohort at diagnosis was 57 yrs (range 32-85) with 75% of them being post menopausal. 93% of the tumors were of the infiltrating ductal carcinoma (IDC) type. Only a quarter of the patients had tumors of less than 2 cm and the majority (70%) of tumors were pT2 or pT3. Of all the patients, the vast majority have received appropriate multi-modal therapy including chemotherapy and endocrine therapy. However, due to financial constraints, only 7% (3/45) of HER2 amplified patients received trastuzumab, and this is in keeping with trends observed at other large tertiary care centers in India [30]. As stated earlier these patients are being followed up and the median follow up as of Jun 30th 2013 is 29 months and median disease specific survival is 27 months.
60% (144/240) of the tumors showed high percentage of ER staining (11-100%), 9% (21/240) showed low percentage of ER staining (1-10%) while 31% (75/240) were negative for ER by IHC. The median age of the low-ER staining group (55 yrs) was 5 years lesser than the high ER immunostaining group and matched with that of the ER negative group. The PgR positivity of the low-ER staining tumors was intermediate between the strongly ER positive group and the ER negative tumors supporting the contention that these tumors are indeed a mixture of the two classes.
There is a good correlation between the ER IHC and ER related gene expression
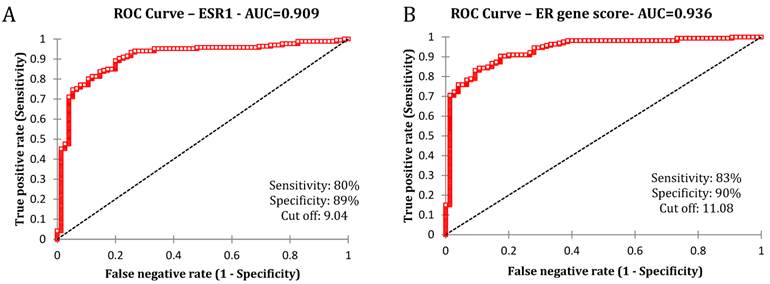
We initially examined the ROC curve of ESR1 gene expression and the ER protein levels estimated by IHC.
The estimated area under the curve showed high concordance (0.909) between ESR1 mRNA and ER protein expression (Fig 1A). We then extended the analysis to the chosen set of luminal genes that have been reported to be highly correlated with the ER positive phenotype. The average expression of ESR1, PgR, GATA3, TFF1, FOXA1 and XBP1 depicted as the "ER gene score" was plotted against the ER expression by IHC. This analysis revealed that the concordance between the extended set of 6 genes and the ER protein remained high (AUC-0.936) suggesting a strong correlation between the two sets of measures (Fig 1B).
Clinical characteristics of the subjects.
| All ( N=235) | ER Groups - N (%) | |||
|---|---|---|---|---|
| N=140 (ER-11-100%) | N=21 (ER-1-10%) | N=74 (ER Neg) | ||
| Age (years) | ||||
| Range | 32-85 | |||
| Mean | 57 | 59 | 54 | 54 |
| Median | 58 | 60 | 55 | 54 |
| Menopausal Status | ||||
| Pre | 58 (24) | 28 (19) | 8 (38) | 22 (30) |
| Post | 177 (76) | 112 (81) | 13 (62) | 52 (70) |
| Total FFPE blocks analyzed | 240 | 144 | 21 | 75 |
| Grade | ||||
| I | 21 (9) | 13 (9) | 3 (14) | 5 (7) |
| II | 105 (43) | 68 (47) | 10 (48) | 27 (36) |
| III | 98 (41) | 54 (38) | 7 (33) | 37 (49) |
| NA | 16 (7) | 9 (6) | 1 (5) | 6 (8) |
| T size | ||||
| <=2 cm | 65 (27) | 44 (30) | 3 (14) | 18 (24) |
| 2.1-5 cm | 138 (58) | 77 (53) | 15 (72) | 46 (62) |
| >5 cm | 29 (12) | 16 (11) | 3 (14) | 10 (13) |
| pTX | 8 (3) | 7 (5) | 0 | 1 (1) |
| Lymph node status | ||||
| Positive | 140 (58) | 88 (61) | 14 (67) | 38 (51) |
| Negative | 91 (38) | 48 (33) | 7 (33) | 36 (48) |
| NA | 9 (4) | 8 (6) | 0 | 1 (1) |
| Stage | ||||
| I | 39 (16) | 23 (16) | 2 (9) | 14 (19) |
| II | 110 (46) | 64 (44) | 9 (43) | 37 (49) |
| III | 87 (36) | 54 (38) | 10 (48) | 23 (31) |
| IV | 4 (2) | 3 (2) | 0 | 1 (1) |
| PR status | ||||
| Positive | 117 (49) | 104 (72) | 9 (43) | 4 (5) |
| Negative | 123 (51) | 40 (28) | 12 (57) | 71 (95) |
| HER2 status and therapy | ||||
| HER2 Positive | 45 (19) | 18 (12) | 5 (24) | 22 (30) |
| HER2 equivocal | 26 (11) | 20 (14) | 0 | 6 (8) |
| HER2 Negative | 169 (70) | 106 (74) | 16 (76) | 47 (62) |
| Trastuzumab treatment | 3 (7) | 1 (5) | 0 | 2 (9) |
| Adjuvant Chemotherapy N=235 | ||||
| Anthracycline and Taxane | 96 (41) | 50 (36) | 10 (48) | 36 (49) |
| Anthracycline plus Other | 50 (21) | 26 (19) | 6 (29) | 18 (24) |
| Other | 15 (6) | 7 (5) | 0 | 8 (11) |
| None | 74 (31) | 57 (41) | 5 (24) | 12 (16) |
| Adjuvant Endocrine therapy | 128 (91) | 15 (71) | ||
Association between ER IHC and chosen genes by the receiver operating characteristics analysis. A) ROC curve between ER IHC and ESR1 mRNA B) ROC curve between ER IHC and average of all genes (ESR1, PgR, GATA3, TFF1, XBP1 and FOXA1)
The majority of low-ER positive tumors are ER negative by the probability score
As explained in methods section, we then used logistic regression models to evaluate the correlation of the expression of each of the chosen genes with the ER status by IHC.
Expression values of the luminal set of genes were used as continuous variable against binomial ER status as positive or negative by IHC. Various combinations of genes used and the BIC (Bayesian Information Criterion) values of the model are shown in Table 3A. The model selected finally (Table 3B.) used ESR1, GATA3 and TFF1 as the best markers for prediction of ER status (Likelihood Ratio test, p<0.001) and calculated the probability score ranging from 0 to 1. From the standardized coefficients we can see that the ESR1 had the highest weight in the composite score, followed by GATA3 and TFF1 as seen in Table 3B.
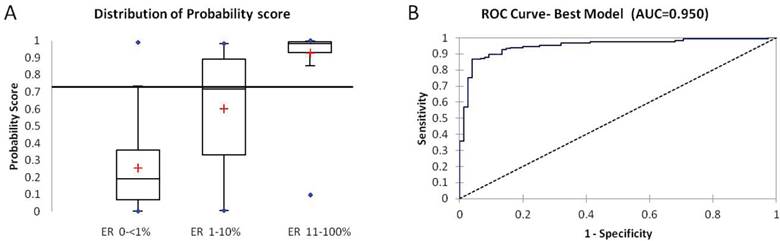
Samples with a probability score above 0.73 were considered ER positive based on the cut off determined by the ROC analysis with ER IHC. Distribution of the probability score between the ER negative (<1%), low ER (1-10%) and ER positive (>11-100%) is shown in Fig 2A. The composite score resulting from the best-fitting model provided a better discriminatory ability than a score based on a simple average of the six genes (AUC increased from 0.93 to 0.95) (Figure. 2B). The mean probability score was highest and tightest in ER 11-100% (0.93), lowest in ER < 1% (0.25) and was 0.6 in the low-ER (1-10%) group (Kruskal-Wallis test, p <0.0001). 95% (71/75) of ER IHC negative and 92% (133/144) of ER IHC positive were classified accurately by the probability score. The tumors in the Low-ER group (1-10%) were almost evenly distributed between the ER positive and negative classes.
Different predictive models compared
| Predictor genes included | Bayesian Information Criterion (BIC) |
|---|---|
| ESR1, GATA3, TFF1 | 150.357 |
| ESR1, GATA3, PgR | 150.726 |
| ESR1, PgR, GATA3, TFF1 | 153.383 |
| ESR1, PgR, GATA3, TFF1 XBP1 | 158.826 |
| ESR1, PgR, GATA3, TFF1, XBP1 and FOXA1 | 162.972 |
Best Fitting Logistic model
| Odds ratio | 95% CI | Standardized Co-efficient | Standard Error | |
|---|---|---|---|---|
| ESR1 | 1.85 | 1.45-2.34 | 0.88 | 0.174 |
| GATA3 | 1.59 | 1.28-1.98 | 0.75 | 0.181 |
| TFF1 | 1.17 | 1.17-1.38 | 0.35 | 0.179 |
Patients with Low-ER positive tumors are intermediate in survival between ER positive and ER negative
Having demonstrated the ability of the gene test with only 3 markers to perform the task of the ER IHC with >90% accuracy we wished to evaluate the clinical validity of the gene expression based probability function by testing its ability to predict prognosis. Samples were divided into low probability and high probability score groups (based on the cutoff of 0.73 determined by ROC analysis with ER IHC) and the survival between the two groups was compared. In the survival analysis, we have used disease specific survival. Though our median follow-up is relatively short we have had outcomes in a significant number of patients (34/235 = 15%) given the larger proportion of later stage disease and the greater proportion of ER negative patients compared to most Western cohorts.
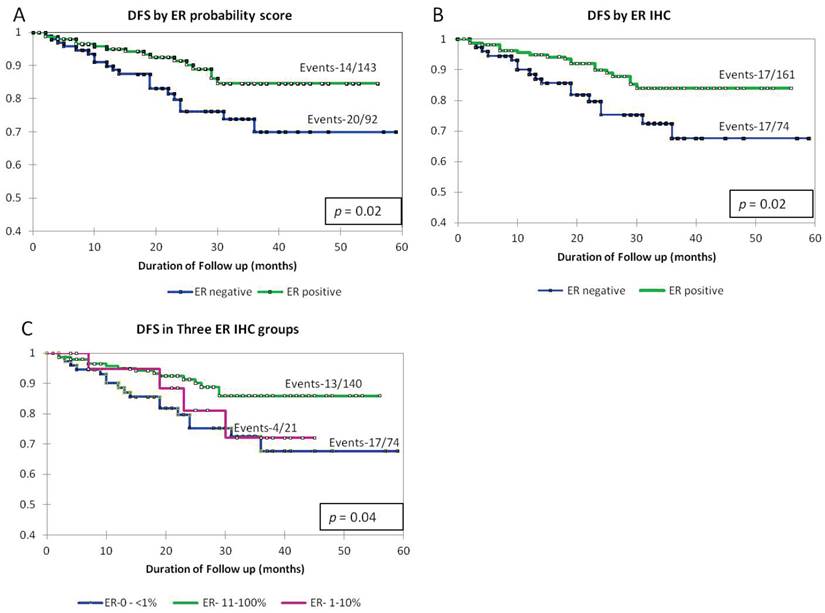
As shown in Fig 3, Kaplan-Meier survival curves in the groups divided by the gene-based probability score were similar to those arrived at by the dichotomizing ER IHC (Fig 3A and B). We then repeated the survival analysis by segregating the low-ER staining group and compared it to the ER >11-100% and ER negative. The survival curve of the low-ER staining group was intermediate between the groups representing high ER positivity and ER negativity (Fig 3C).
A: Box plot of distribution of probability score from the best fitting logistic regression model within the three IHC defined groups (ER 0-<1%, 1-10%, 11-100%). B: ROC Curve based on the best fitting model improves AUC (0.95).
Kaplan-Meier survival curves showing disease specific survival by A) dichotomizing the samples into ER positive and negative (cut off of 0.73) by ER probability score as generated by the model, B) by ER IHC as negative (<1% staining) and positive (>1% staining), C) in three groups defined by ER IHC as ER <1%, 1-10% and 11-100%
Discussion
Undoubtedly the ER protein is the marker with the greatest proven clinical utility as a predictive marker of response to hormonal therapy in breast cancer patients. In addition it has been shown to be strongly correlated with a more favorable prognosis. In conjunction with other clinico-pathological variables that include the menopausal status, lymph node status, tumor size, tumor grade, PgR and HER2, it has been used in prognostic indices like Adjuvant! Online [31] The classic work of Perou et. al.,[10] and Sorlie et. al.,[32] in the early 2000s paved the way for the use of gene expression patterns to study the diversity of these cancers. The first major diagnostic advance for the subset of ER positive, node negative patients came through the introduction of OncotypeDX, which is based on the estimation of the transcript abundances of a set of 16 test genes including ESR1, PgR, HER2 using FFPE specimens[16]. The more general principle that emerged from the development of these approaches was the establishment of strong concordance between transcript and protein levels and the support for using multiple markers to achieve better predictability of a particular phenotypic trait as suggested by Allred [33]. Oncotype DX also opened the doors for the use of FFPE specimens for obtaining gene expression signatures of clinical utility.
The decrease in threshold for determining ER positivity by IHC from the previous level of 10% to 1% by ASCO in 2010 has created a group of low ER positive tumors with the nuclear staining of ER ranging from 1-9%[9]. These tumors would have been considered negative using the earlier cut-off. We have examined the biology of these tumors using multiple markers rather than just ESR1 alone based on the demonstration that multiple related markers tend to improve the predictive performance of single markers.
The markers we have used encompass both direct transcriptional regulators of ESR1 expression as well as genes that need the presence of ER protein for their sustained expression [11-14]. The categorization of approx half (48%) of the low-ER tumors as belonging to the ER negative group by the use of the gene-expression based probability model suggests the absence of functional ER activity in these tumors. Survival analysis of this group was seen to be intermediate between those of ER positive (11-100%) and ER negative (<1%) further reiterating the fact of them being a mixed group of tumors as shown by our model.
The selected logistic regression model included only three (ESR1, GATA3 and TFF1) out of the 6 genes considered by us to be significant predictors of ER status. The GATA3 and TFF1 genes have been shown to be closely regulated by ER through cross loop mechanisms [34]. We have used a receiver operating characteristic curve to determine the cut-off with highest specificity and sensitivity in the probability score against ER IHC as the gold standard. At this cut-off, the agreement between the transcript-based probability score and ER IHC status was higher among ER IHC negatives (95%) than among ER IHC positives (92%). Given many of the low-ER tumors behave as ER negative tumors, it is not surprising to see a greater disagreement between the two methods in the ER IHC positive category. Though ER IHC is the current gold-standard for clinical diagnosis of ER positivity or for the evaluation of any new ER assay, it may not be ideal. Future research could consider the use of more sophisticated statistical models, such as latent class models, to determine the correlation between the transcript-based score and the true ER status [35].
Two other studies published recently [20, 21] have also tested the molecular phenotypes of low ER staining tumors. Iwamoto et. al., compared the ESR1 mRNA and a 106 probe-set ER associated gene-signature by microarray analysis in 31 low-ER staining (1-10%) and 251 ER positive (>10%) tumors. ESR1 mRNA expression levels in their results were significantly different between the > 10% and 1-9% ER staining tumors. PAM 50 subtype classification showed that only a small proportion (16%) of the low-ER was Luminal, and 42% were Basal-like and other subtypes. Similarly, Deyarmin et. al., determined the intrinsic subtype of the low-ER staining tumors (n=26) by microarray analysis and showed 88% of them were either Basal-like or HER2 Enriched. Only tumors with staining of 10% could be classified as Luminal subtype. Our findings are in strong agreement with both these studies in that the majority of tumors with low-ER show clinico-pathological and molecular profiles like ER negative tumors.
A majority of the clinically HER2 positive patients (93% in our study; 91% at the Tata Memorial Hospital, Mumbai) in India do not receive trastuzumab due to financial constraints. To exclude the possibility that the difference between the survival curves might be the effect of clinical HER2 positive patient not receiving trastuzumab, we performed the analysis of survival after eliminating the HER2 positive patients from both groups, and this did not alter the separation of the survival curves (data not shown). This suggests that the difference in survival between the ER positive and ER negative patients is not skewed by HER2 status or treatment.
The observations of this report while of significant interest, need to be confirmed in a study with larger numbers, since we had only 21 patients in the border-line ER positive category. Efforts are underway to test patient specimens from an independent cohort from a local cancer centre. In addition, since the ASCO guidelines for ER positivity changed midway during the study, it is important that these conclusions are tested in a setting wherein the 1% cut-off is used prospectively for all treatment decisions. Only then will it be ready for use in routine clinical practice.
The molecular testing in our study was performed by SYBR Green qPCR of just 3 markers using nucleic acids extracted from FFPE sections contiguous to the ones on which IHC was performed. One of the implications of our study is that in the future the adoption of multiple markers for determining the hormone receptor status rather than ER and PgR IHC alone might improve the certainty of categorization. We believe that the simplicity of the methods and the limited number of markers used in our testing will enable easier transfer to the clinical lab when it is ready for use.
Abbreviations
ER: Estrogen Receptor; IHC: Immunohistochemistry; FFPE: Formalin Fixed and Paraffin-Embedded; ASCO: American Society of Clinical Oncology; FISH: Fluorescent in situ Hybridization; HKG: House Keeping Gene; RNA: Ribonucleic Acid; q-RT-PCR: Quantitative Reverse Transcriptase Polymerase Chain Reaction; cDNA: complementary Deoxyribonucleic Acid; Ct: Cycle of threshold; RNU: Relative Normalized Units; ROC: Receiver Operating Charactersitics; BIC: Bayesian Information Criterion; IDC: Infiltrating Ductal Carcinoma: DFS: Disease Specific Survival.
Acknowledgements
This work was funded by the Nadathur Holdings, Bangalore and The Bagaria Education Trust, Bangalore, India. Author Nandini Dendukuri is supported by a Chercheur Boursier Senior award from the Fonds de Recherche Québec - Santé.
Competing Interests
The authors have declared that no competing interest exists.
References
1. DeSombre ER, Smith S, Block GE. et al. Prediction of breast cancer response to endocrine therapy. Cancer chemotherapy reports Part 1. 1974;58:513-9
2. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival. an overview of the randomised trials. The Lancet. 2005;365:1687-717 doi:10.1016/s0140-6736(05)66544-0
3. Allred DC, Bustamante MA, Daniel CO. et al. Immunocytochemical analysis of estrogen receptors in human breast carcinomas. Evaluation of 130 cases and review of the literature regarding concordance with biochemical assay and clinical relevance. Archives of surgery (Chicago, Ill: 1960). 1990;125:107-13
4. Elledge RM, Green S, Pugh R. et al. Estrogen receptor (ER) and progesterone receptor (PgR), by ligand-binding assay compared with ER, PgR and pS2, by immuno-histochemistry in predicting response to tamoxifen in metastatic breast cancer: a Southwest Oncology Group Study. International journal of cancer Journal international du cancer. 2000;89:111-7
5. Badve S, Vladislav IT, Spaulding B. et al. EP1: a novel rabbit monoclonal antibody for detection of oestrogen receptor alpha. Journal of clinical pathology. 2013 doi:10.1136/jclinpath-2012-201391
6. Kim C, Tang G, Pogue-Geile KL. et al. Estrogen receptor (ESR1) mRNA expression and benefit from tamoxifen in the treatment and prevention of estrogen receptor-positive breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29:4160-7 doi:10.1200/jco.2010.32.9615
7. Denkert C, Loibl S, Kronenwett R. et al. RNA-based determination of ESR1 and HER2 expression and response to neoadjuvant chemotherapy. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 2013;24:632-9 doi:10.1093/annonc/mds339
8. Harvey JM, Clark GM, Osborne CK. et al. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 1999;17:1474-81
9. Hammond ME, Hayes DF, Dowsett M. et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28:2784-95 doi:10.1200/JCO.2009.25.6529
10. Perou CM, Sorlie T, Eisen MB. et al. Molecular portraits of human breast tumours. Nature. 2000;406:747-52 doi:10.1038/35021093
11. Gruvberger S, Ringner M, Chen Y. et al. Estrogen receptor status in breast cancer is associated with remarkably distinct gene expression patterns. Cancer research. 2001;61:5979-84
12. Schneider J, Ruschhaupt M, Buness A. et al. Identification and meta-analysis of a small gene expression signature for the diagnosis of estrogen receptor status in invasive ductal breast cancer. International journal of cancer Journal international du cancer. 2006;119:2974-9 doi:10.1002/ijc.22234
13. Badve S, Turbin D, Thorat MA. et al. FOXA1 expression in breast cancer--correlation with luminal subtype A and survival. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13:4415-21 doi:10.1158/1078-0432.CCR-07-0122
14. Symmans WF, Hatzis C, Sotiriou C. et al. Genomic index of sensitivity to endocrine therapy for breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28:4111-9 doi:10.1200/JCO.2010.28.4273
15. van 't Veer LJ, Dai H, van de Vijver MJ. et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530-6 doi:10.1038/415530a
16. Paik S, Shak S, Tang G. et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. The New England journal of medicine. 2004;351:2817-26 doi:10.1056/NEJMoa041588
17. Badve SS, Baehner FL, Gray RP. et al. Estrogen- and progesterone-receptor status in ECOG 2197: comparison of immunohistochemistry by local and central laboratories and quantitative reverse transcription polymerase chain reaction by central laboratory. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2008;26:2473-81 doi:10.1200/JCO.2007.13.6424
18. Harbeck N, Rody A. Lost in translation? Estrogen receptor status and endocrine responsiveness in breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2012;30:686-9 doi:10.1200/jco.2011.38.9619
19. Wolff AC, Dowsett M. Estrogen receptor: a never ending story? Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29:2955-8 doi:10.1200/jco.2011.35.4589
20. Iwamoto T, Booser D, Valero V. et al. Estrogen receptor (ER) mRNA and ER-related gene expression in breast cancers that are 1% to 10% ER-positive by immunohistochemistry. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2012;30:729-34 doi:10.1200/JCO.2011.36.2574
21. Deyarmin B, Kane JL, Valente AL. et al. Effect of ASCO/CAP guidelines for determining ER status on molecular subtype. Annals of surgical oncology. 2013;20:87-93 doi:10.1245/s10434-012-2588-8
22. Desai SB, Moonim MT, Gill AK. et al. Hormone receptor status of breast cancer in India: a study of 798 tumours. Breast. 2000;9:267-70 discussion 70. doi:10.1054/brst.2000.0134
23. Green M, Raina V. Epidemiology, screening and diagnosis of breast cancer in the Asia-Pacific region: Current perspectives and important considerations. Asia-Pacific Journal of Clinical Oncology. 2008;4:S5-S13 doi:10.1111/j.1743-7563.2008.00191.x
24. Shet T, Agrawal A, Nadkarni M. et al. Hormone receptors over the last 8 years in a cancer referral center in India: What was and what is? Indian Journal of Pathology and Microbiology. 2009;52:171. doi:10.4103/0377-4929.48909
25. Dey S, Boffetta P, Mathews A. et al. Risk factors according to estrogen receptor status of breast cancer patients in Trivandrum, South India. International journal of cancer Journal international du cancer. 2009;125:1663-70 doi:10.1002/ijc.24460
26. Manjunath S, Prabhu JS, Kaluve R. et al. Estrogen Receptor Negative Breast Cancer in India: Do We Really Have Higher Burden of this Subtype? Indian journal of surgical oncology. 2011;2:122-5 doi:10.1007/s13193-011-0072-8
27. Leong SP, Shen ZZ, Liu TJ. et al. Is breast cancer the same disease in Asian and Western countries? World journal of surgery. 2010;34:2308-24 doi:10.1007/s00268-010-0683-1
28. Korlimarla A, Prabhu JS, Anupama CE. et al. Separate Quality-Control Measures Are Necessary for Estimation of RNA and Methylated DNA from Formalin-Fixed, Paraffin-Embedded Specimens by Quantitative PCR. The Journal of molecular diagnostics. 2014 In press
29. Prabhu JS, Wahi K, Korlimarla A. et al. The epigenetic silencing of the estrogen receptor (ER) by hypermethylation of the ESR1 promoter is seen predominantly in triple-negative breast cancers in Indian women. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2012;33:315-23 doi:10.1007/s13277-012-0343-1
30. Ghosh J, Gupta S, Desai S. et al. Estrogen, progesterone and HER2 receptor expression in breast tumors of patients, and their usage of HER2-targeted therapy, in a tertiary care centre in India. Indian journal of cancer. 2011;48:391-6 doi:10.4103/0019-509X.92245
31. Ravdin PM, Siminoff LA, Davis GJ. et al. Computer program to assist in making decisions about adjuvant therapy for women with early breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2001;19:980-91
32. Sorlie T, Perou CM, Tibshirani R. et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10869-74 doi:10.1073/pnas.191367098
33. Allred DC. Problems and solutions in the evaluation of hormone receptors in breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2008;26:2433-5 doi:10.1200/JCO.2007.15.7800
34. Hoch RV, Thompson DA, Baker RJ. et al. GATA-3 is expressed in association with estrogen receptor in breast cancer. International journal of cancer Journal international du cancer. 1999;84:122-8
35. Skrondal A, Rabe-Hesketh S. Generalized latent variable modeling. Multilevel, longitudinal, and structural equation models. CRC Press. 2004
Author contact
![]() Corresponding author: Jyothi S Prabhu. Fax No.080-25532037; Email: jyothires.in; jyothisprabhucom
Corresponding author: Jyothi S Prabhu. Fax No.080-25532037; Email: jyothires.in; jyothisprabhucom

 Global reach, higher impact
Global reach, higher impact